Jacinto C. Nascimento
DALL-M: Context-Aware Clinical Data Augmentation with LLMs
Jul 11, 2024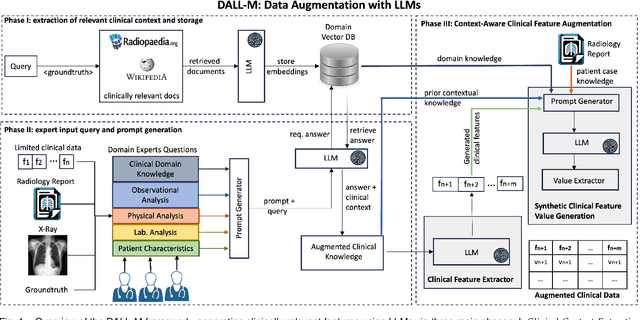


Abstract:X-ray images are vital in medical diagnostics, but their effectiveness is limited without clinical context. Radiologists often find chest X-rays insufficient for diagnosing underlying diseases, necessitating comprehensive clinical features and data integration. We present a novel technique to enhance the clinical context through augmentation techniques with clinical tabular data, thereby improving its applicability and reliability in AI medical diagnostics. To address this, we introduce a pioneering approach to clinical data augmentation that employs large language models (LLMs) to generate patient contextual synthetic data. This methodology is crucial for training more robust deep learning models in healthcare. It preserves the integrity of real patient data while enriching the dataset with contextually relevant synthetic features, significantly enhancing model performance. DALL-M uses a three-phase feature generation process: (i) clinical context storage, (ii) expert query generation, and (iii) context-aware feature augmentation. DALL-M generates new, clinically relevant features by synthesizing chest X-ray images and reports. Applied to 799 cases using nine features from the MIMIC-IV dataset, it created an augmented set of 91 features. This is the first work to generate contextual values for existing and new features based on patients' X-ray reports, gender, and age and to produce new contextual knowledge during data augmentation. Empirical validation with machine learning models, including Decision Trees, Random Forests, XGBoost, and TabNET, showed significant performance improvements. Incorporating augmented features increased the F1 score by 16.5% and Precision and Recall by approximately 25%. DALL-M addresses a critical gap in clinical data augmentation, offering a robust framework for generating contextually enriched datasets.
Correspondence Free Multivector Cloud Registration using Conformal Geometric Algebra
Jun 17, 2024Abstract:We present, for the first time, a novel theoretical approach to address the problem of correspondence free multivector cloud registration in conformal geometric algebra. Such formalism achieves several favorable properties. Primarily, it forms an orthogonal automorphism that extends beyond the typical vector space to the entire conformal geometric algebra while respecting the multivector grading. Concretely, the registration can be viewed as an orthogonal transformation (\it i.e., scale, translation, rotation) belonging to $SO(4,1)$ - group of special orthogonal transformations in conformal geometric algebra. We will show that such formalism is able to: $(i)$ perform the registration without directly accessing the input multivectors. Instead, we use primitives or geometric objects provided by the conformal model - the multivectors, $(ii)$ the geometric objects are obtained by solving a multilinear eigenvalue problem to find sets of eigenmultivectors. In this way, we can explicitly avoid solving the correspondences in the registration process. Most importantly, this offers rotation and translation equivariant properties between the input multivectors and the eigenmultivectors. Experimental evaluation is conducted in datasets commonly used in point cloud registration, to testify the usefulness of the approach with emphasis to ambiguities arising from high levels of noise. The code is available at https://github.com/Numerical-Geometric-Algebra/RegistrationGA . This work was submitted to the International Journal of Computer Vision and is currently under review.
SelfReDepth: Self-Supervised Real-Time Depth Restoration for Consumer-Grade Sensors
Jun 05, 2024Abstract:Depth maps produced by consumer-grade sensors suffer from inaccurate measurements and missing data from either system or scene-specific sources. Data-driven denoising algorithms can mitigate such problems. However, they require vast amounts of ground truth depth data. Recent research has tackled this limitation using self-supervised learning techniques, but it requires multiple RGB-D sensors. Moreover, most existing approaches focus on denoising single isolated depth maps or specific subjects of interest, highlighting a need for methods to effectively denoise depth maps in real-time dynamic environments. This paper extends state-of-the-art approaches for depth-denoising commodity depth devices, proposing SelfReDepth, a self-supervised deep learning technique for depth restoration, via denoising and hole-filling by inpainting full-depth maps captured with RGB-D sensors. The algorithm targets depth data in video streams, utilizing multiple sequential depth frames coupled with color data to achieve high-quality depth videos with temporal coherence. Finally, SelfReDepth is designed to be compatible with various RGB-D sensors and usable in real-time scenarios as a pre-processing step before applying other depth-dependent algorithms. Our results demonstrate our approach's real-time performance on real-world datasets. They show that it outperforms state-of-the-art denoising and restoration performance at over 30fps on Commercial Depth Cameras, with potential benefits for augmented and mixed-reality applications.
* 13pp, 5 figures, 1 table
Key Patches Are All You Need: A Multiple Instance Learning Framework For Robust Medical Diagnosis
May 02, 2024Abstract:Deep learning models have revolutionized the field of medical image analysis, due to their outstanding performances. However, they are sensitive to spurious correlations, often taking advantage of dataset bias to improve results for in-domain data, but jeopardizing their generalization capabilities. In this paper, we propose to limit the amount of information these models use to reach the final classification, by using a multiple instance learning (MIL) framework. MIL forces the model to use only a (small) subset of patches in the image, identifying discriminative regions. This mimics the clinical procedures, where medical decisions are based on localized findings. We evaluate our framework on two medical applications: skin cancer diagnosis using dermoscopy and breast cancer diagnosis using mammography. Our results show that using only a subset of the patches does not compromise diagnostic performance for in-domain data, compared to the baseline approaches. However, our approach is more robust to shifts in patient demographics, while also providing more detailed explanations about which regions contributed to the decision. Code is available at: https://github.com/diogojpa99/MedicalMultiple-Instance-Learning.
Latent Embedding Clustering for Occlusion Robust Head Pose Estimation
Mar 29, 2024Abstract:Head pose estimation has become a crucial area of research in computer vision given its usefulness in a wide range of applications, including robotics, surveillance, or driver attention monitoring. One of the most difficult challenges in this field is managing head occlusions that frequently take place in real-world scenarios. In this paper, we propose a novel and efficient framework that is robust in real world head occlusion scenarios. In particular, we propose an unsupervised latent embedding clustering with regression and classification components for each pose angle. The model optimizes latent feature representations for occluded and non-occluded images through a clustering term while improving fine-grained angle predictions. Experimental evaluation on in-the-wild head pose benchmark datasets reveal competitive performance in comparison to state-of-the-art methodologies with the advantage of having a significant data reduction. We observe a substantial improvement in occluded head pose estimation. Also, an ablation study is conducted to ascertain the impact of the clustering term within our proposed framework.
2D Image head pose estimation via latent space regression under occlusion settings
Nov 10, 2023Abstract:Head orientation is a challenging Computer Vision problem that has been extensively researched having a wide variety of applications. However, current state-of-the-art systems still underperform in the presence of occlusions and are unreliable for many task applications in such scenarios. This work proposes a novel deep learning approach for the problem of head pose estimation under occlusions. The strategy is based on latent space regression as a fundamental key to better structure the problem for occluded scenarios. Our model surpasses several state-of-the-art methodologies for occluded HPE, and achieves similar accuracy for non-occluded scenarios. We demonstrate the usefulness of the proposed approach with: (i) two synthetically occluded versions of the BIWI and AFLW2000 datasets, (ii) real-life occlusions of the Pandora dataset, and (iii) a real-life application to human-robot interaction scenarios where face occlusions often occur. Specifically, the autonomous feeding from a robotic arm.
MDF-Net: Multimodal Dual-Fusion Network for Abnormality Detection using CXR Images and Clinical Data
Feb 26, 2023Abstract:This study aims to investigate the effects of including patients' clinical information on the performance of deep learning (DL) classifiers for disease location in chest X-ray images. Although current classifiers achieve high performance using chest X-ray images alone, our interviews with radiologists indicate that clinical data is highly informative and essential for interpreting images and making proper diagnoses. In this work, we propose a novel architecture consisting of two fusion methods that enable the model to simultaneously process patients' clinical data (structured data) and chest X-rays (image data). Since these data modalities are in different dimensional spaces, we propose a spatial arrangement strategy, termed spatialization, to facilitate the multimodal learning process in a Mask R-CNN model. We performed an extensive experimental evaluation comprising three datasets with different modalities: MIMIC CXR (chest X-ray images), MIMIC IV-ED (patients' clinical data), and REFLACX (annotations of disease locations in chest X-rays). Results show that incorporating patients' clinical data in a DL model together with the proposed fusion methods improves the performance of disease localization in chest X-rays by 12\% in terms of Average Precision compared to a standard Mask R-CNN using only chest X-rays. Further ablation studies also emphasize the importance of multimodal DL architectures and the incorporation of patients' clinical data in disease localisation. The architecture proposed in this work is publicly available to promote the scientific reproducibility of our study (https://github.com/ChihchengHsieh/multimodal-abnormalities-detection).
Censor-aware Semi-supervised Learning for Survival Time Prediction from Medical Images
May 26, 2022



Abstract:Survival time prediction from medical images is important for treatment planning, where accurate estimations can improve healthcare quality. One issue affecting the training of survival models is censored data. Most of the current survival prediction approaches are based on Cox models that can deal with censored data, but their application scope is limited because they output a hazard function instead of a survival time. On the other hand, methods that predict survival time usually ignore censored data, resulting in an under-utilization of the training set. In this work, we propose a new training method that predicts survival time using all censored and uncensored data. We propose to treat censored data as samples with a lower-bound time to death and estimate pseudo labels to semi-supervise a censor-aware survival time regressor. We evaluate our method on pathology and x-ray images from the TCGA-GM and NLST datasets. Our results establish the state-of-the-art survival prediction accuracy on both datasets.
Post-hoc Overall Survival Time Prediction from Brain MRI
Feb 22, 2021



Abstract:Overall survival (OS) time prediction is one of the most common estimates of the prognosis of gliomas and is used to design an appropriate treatment planning. State-of-the-art (SOTA) methods for OS time prediction follow a pre-hoc approach that require computing the segmentation map of the glioma tumor sub-regions (necrotic, edema tumor, enhancing tumor) for estimating OS time. However, the training of the segmentation methods require ground truth segmentation labels which are tedious and expensive to obtain. Given that most of the large-scale data sets available from hospitals are unlikely to contain such precise segmentation, those SOTA methods have limited applicability. In this paper, we introduce a new post-hoc method for OS time prediction that does not require segmentation map annotation for training. Our model uses medical image and patient demographics (represented by age) as inputs to estimate the OS time and to estimate a saliency map that localizes the tumor as a way to explain the OS time prediction in a post-hoc manner. It is worth emphasizing that although our model can localize tumors, it uses only the ground truth OS time as training signal, i.e., no segmentation labels are needed. We evaluate our post-hoc method on the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2019 data set and show that it achieves competitive results compared to pre-hoc methods with the advantage of not requiring segmentation labels for training.
Region Proposals for Saliency Map Refinement for Weakly-supervised Disease Localisation and Classification
May 22, 2020



Abstract:The deployment of automated systems to diagnose diseases from medical images is challenged by the requirement to localise the diagnosed diseases to justify or explain the classification decision. This requirement is hard to fulfil because most of the training sets available to develop these systems only contain global annotations, making the localisation of diseases a weakly supervised approach. The main methods designed for weakly supervised disease classification and localisation rely on saliency or attention maps that are not specifically trained for localisation, or on region proposals that can not be refined to produce accurate detections. In this paper, we introduce a new model that combines region proposal and saliency detection to overcome both limitations for weakly supervised disease classification and localisation. Using the ChestX-ray14 data set, we show that our proposed model establishes the new state-of-the-art for weakly-supervised disease diagnosis and localisation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge