Alceu Bissoto
Label-free estimation of clinically relevant performance metrics under distribution shifts
Jul 30, 2025Abstract:Performance monitoring is essential for safe clinical deployment of image classification models. However, because ground-truth labels are typically unavailable in the target dataset, direct assessment of real-world model performance is infeasible. State-of-the-art performance estimation methods address this by leveraging confidence scores to estimate the target accuracy. Despite being a promising direction, the established methods mainly estimate the model's accuracy and are rarely evaluated in a clinical domain, where strong class imbalances and dataset shifts are common. Our contributions are twofold: First, we introduce generalisations of existing performance prediction methods that directly estimate the full confusion matrix. Then, we benchmark their performance on chest x-ray data in real-world distribution shifts as well as simulated covariate and prevalence shifts. The proposed confusion matrix estimation methods reliably predicted clinically relevant counting metrics on medical images under distribution shifts. However, our simulated shift scenarios exposed important failure modes of current performance estimation techniques, calling for a better understanding of real-world deployment contexts when implementing these performance monitoring techniques for postmarket surveillance of medical AI models.
A Real-Time Digital Twin for Type 1 Diabetes using Simulation-Based Inference
Jul 02, 2025Abstract:Accurately estimating parameters of physiological models is essential to achieving reliable digital twins. For Type 1 Diabetes, this is particularly challenging due to the complexity of glucose-insulin interactions. Traditional methods based on Markov Chain Monte Carlo struggle with high-dimensional parameter spaces and fit parameters from scratch at inference time, making them slow and computationally expensive. In this study, we propose a Simulation-Based Inference approach based on Neural Posterior Estimation to efficiently capture the complex relationships between meal intake, insulin, and glucose level, providing faster, amortized inference. Our experiments demonstrate that SBI not only outperforms traditional methods in parameter estimation but also generalizes better to unseen conditions, offering real-time posterior inference with reliable uncertainty quantification.
Subgroup Performance Analysis in Hidden Stratifications
Mar 13, 2025Abstract:Machine learning (ML) models may suffer from significant performance disparities between patient groups. Identifying such disparities by monitoring performance at a granular level is crucial for safely deploying ML to each patient. Traditional subgroup analysis based on metadata can expose performance disparities only if the available metadata (e.g., patient sex) sufficiently reflects the main reasons for performance variability, which is not common. Subgroup discovery techniques that identify cohesive subgroups based on learned feature representations appear as a potential solution: They could expose hidden stratifications and provide more granular subgroup performance reports. However, subgroup discovery is challenging to evaluate even as a standalone task, as ground truth stratification labels do not exist in real data. Subgroup discovery has thus neither been applied nor evaluated for the application of subgroup performance monitoring. Here, we apply subgroup discovery for performance monitoring in chest x-ray and skin lesion classification. We propose novel evaluation strategies and show that a simplified subgroup discovery method without access to classification labels or metadata can expose larger performance disparities than traditional metadata-based subgroup analysis. We provide the first compelling evidence that subgroup discovery can serve as an important tool for comprehensive performance validation and monitoring of trustworthy AI in medicine.
Are Explanations Helpful? A Comparative Analysis of Explainability Methods in Skin Lesion Classifiers
Dec 04, 2024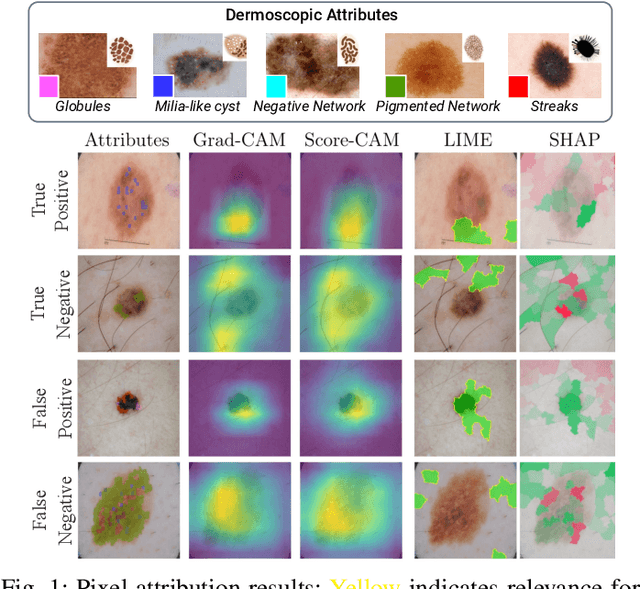
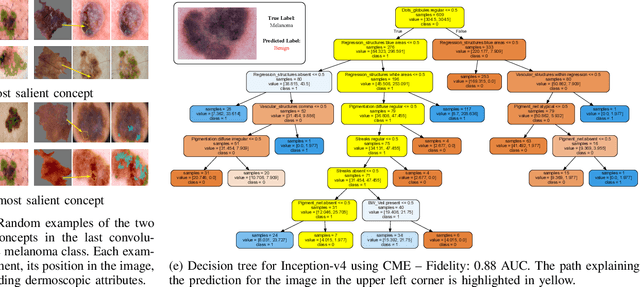
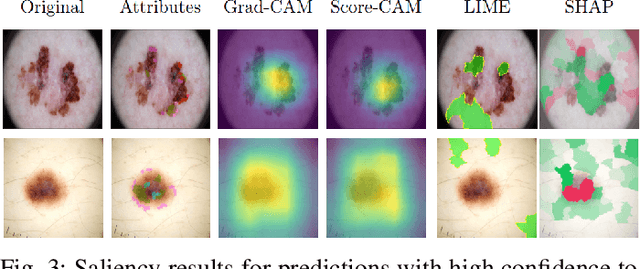
Abstract:Deep Learning has shown outstanding results in computer vision tasks; healthcare is no exception. However, there is no straightforward way to expose the decision-making process of DL models. Good accuracy is not enough for skin cancer predictions. Understanding the model's behavior is crucial for clinical application and reliable outcomes. In this work, we identify desiderata for explanations in skin-lesion models. We analyzed seven methods, four based on pixel-attribution (Grad-CAM, Score-CAM, LIME, SHAP) and three on high-level concepts (ACE, ICE, CME), for a deep neural network trained on the International Skin Imaging Collaboration Archive. Our findings indicate that while these techniques reveal biases, there is room for improving the comprehensiveness of explanations to achieve transparency in skin-lesion models.
BiasPruner: Debiased Continual Learning for Medical Image Classification
Jul 11, 2024Abstract:Continual Learning (CL) is crucial for enabling networks to dynamically adapt as they learn new tasks sequentially, accommodating new data and classes without catastrophic forgetting. Diverging from conventional perspectives on CL, our paper introduces a new perspective wherein forgetting could actually benefit the sequential learning paradigm. Specifically, we present BiasPruner, a CL framework that intentionally forgets spurious correlations in the training data that could lead to shortcut learning. Utilizing a new bias score that measures the contribution of each unit in the network to learning spurious features, BiasPruner prunes those units with the highest bias scores to form a debiased subnetwork preserved for a given task. As BiasPruner learns a new task, it constructs a new debiased subnetwork, potentially incorporating units from previous subnetworks, which improves adaptation and performance on the new task. During inference, BiasPruner employs a simple task-agnostic approach to select the best debiased subnetwork for predictions. We conduct experiments on three medical datasets for skin lesion classification and chest X-Ray classification and demonstrate that BiasPruner consistently outperforms SOTA CL methods in terms of classification performance and fairness. Our code is available here.
Back to the Basics on Predicting Transfer Performance
May 30, 2024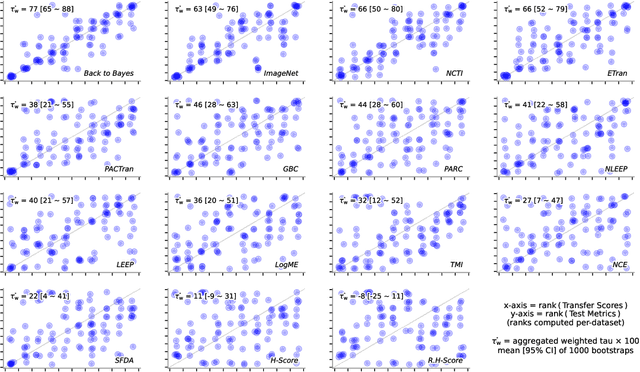
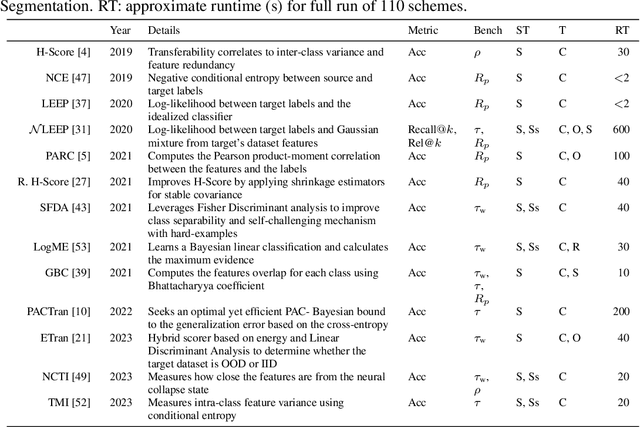
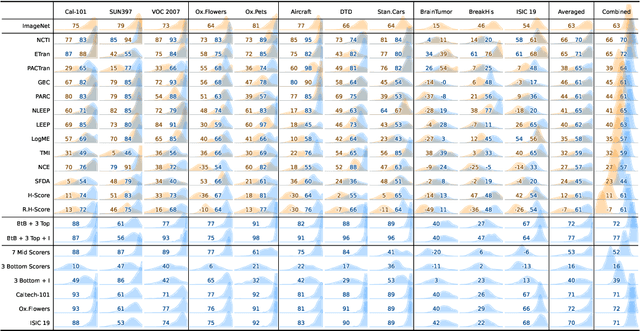
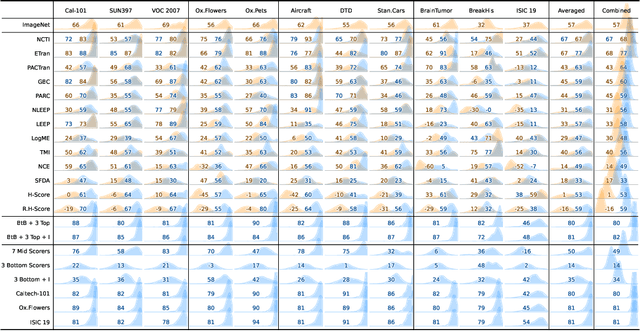
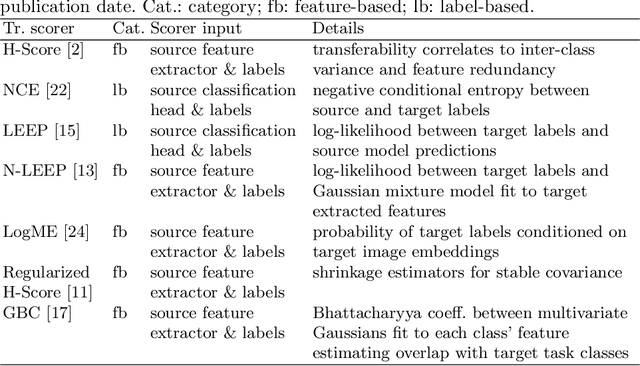
Abstract:In the evolving landscape of deep learning, selecting the best pre-trained models from a growing number of choices is a challenge. Transferability scorers propose alleviating this scenario, but their recent proliferation, ironically, poses the challenge of their own assessment. In this work, we propose both robust benchmark guidelines for transferability scorers, and a well-founded technique to combine multiple scorers, which we show consistently improves their results. We extensively evaluate 13 scorers from literature across 11 datasets, comprising generalist, fine-grained, and medical imaging datasets. We show that few scorers match the predictive performance of the simple raw metric of models on ImageNet, and that all predictors suffer on medical datasets. Our results highlight the potential of combining different information sources for reliably predicting transferability across varied domains.
Key Patches Are All You Need: A Multiple Instance Learning Framework For Robust Medical Diagnosis
May 02, 2024Abstract:Deep learning models have revolutionized the field of medical image analysis, due to their outstanding performances. However, they are sensitive to spurious correlations, often taking advantage of dataset bias to improve results for in-domain data, but jeopardizing their generalization capabilities. In this paper, we propose to limit the amount of information these models use to reach the final classification, by using a multiple instance learning (MIL) framework. MIL forces the model to use only a (small) subset of patches in the image, identifying discriminative regions. This mimics the clinical procedures, where medical decisions are based on localized findings. We evaluate our framework on two medical applications: skin cancer diagnosis using dermoscopy and breast cancer diagnosis using mammography. Our results show that using only a subset of the patches does not compromise diagnostic performance for in-domain data, compared to the baseline approaches. However, our approach is more robust to shifts in patient demographics, while also providing more detailed explanations about which regions contributed to the decision. Code is available at: https://github.com/diogojpa99/MedicalMultiple-Instance-Learning.
MMIST-ccRCC: A Real World Medical Dataset for the Development of Multi-Modal Systems
May 02, 2024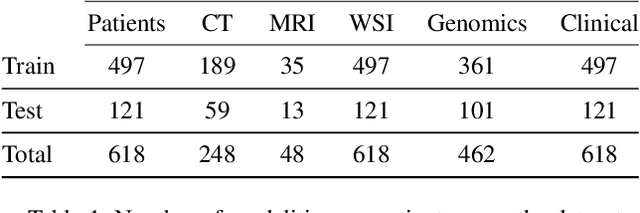
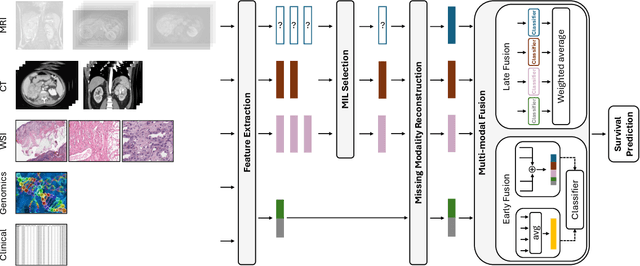
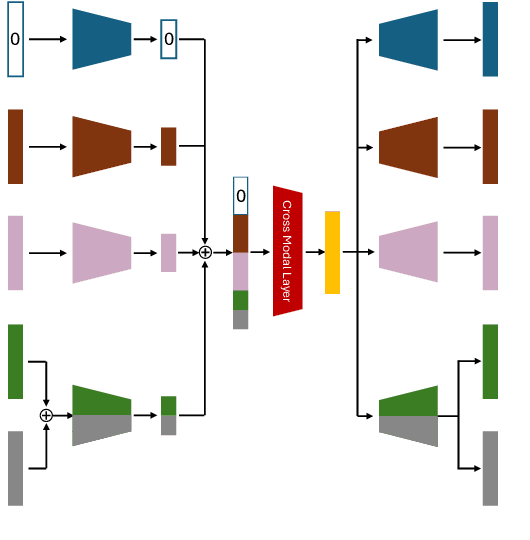
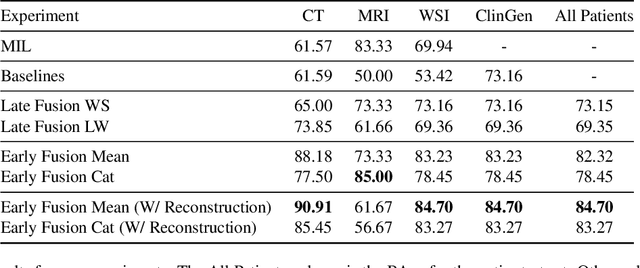
Abstract:The acquisition of different data modalities can enhance our knowledge and understanding of various diseases, paving the way for a more personalized healthcare. Thus, medicine is progressively moving towards the generation of massive amounts of multi-modal data (\emph{e.g,} molecular, radiology, and histopathology). While this may seem like an ideal environment to capitalize data-centric machine learning approaches, most methods still focus on exploring a single or a pair of modalities due to a variety of reasons: i) lack of ready to use curated datasets; ii) difficulty in identifying the best multi-modal fusion strategy; and iii) missing modalities across patients. In this paper we introduce a real world multi-modal dataset called MMIST-CCRCC that comprises 2 radiology modalities (CT and MRI), histopathology, genomics, and clinical data from 618 patients with clear cell renal cell carcinoma (ccRCC). We provide single and multi-modal (early and late fusion) benchmarks in the task of 12-month survival prediction in the challenging scenario of one or more missing modalities for each patient, with missing rates that range from 26$\%$ for genomics data to more than 90$\%$ for MRI. We show that even with such severe missing rates the fusion of modalities leads to improvements in the survival forecasting. Additionally, incorporating a strategy to generate the latent representations of the missing modalities given the available ones further improves the performance, highlighting a potential complementarity across modalities. Our dataset and code are available here: https://multi-modal-ist.github.io/datasets/ccRCC
XAI for Skin Cancer Detection with Prototypes and Non-Expert Supervision
Feb 02, 2024Abstract:Skin cancer detection through dermoscopy image analysis is a critical task. However, existing models used for this purpose often lack interpretability and reliability, raising the concern of physicians due to their black-box nature. In this paper, we propose a novel approach for the diagnosis of melanoma using an interpretable prototypical-part model. We introduce a guided supervision based on non-expert feedback through the incorporation of: 1) binary masks, obtained automatically using a segmentation network; and 2) user-refined prototypes. These two distinct information pathways aim to ensure that the learned prototypes correspond to relevant areas within the skin lesion, excluding confounding factors beyond its boundaries. Experimental results demonstrate that, even without expert supervision, our approach achieves superior performance and generalization compared to non-interpretable models.
The Performance of Transferability Metrics does not Translate to Medical Tasks
Aug 14, 2023

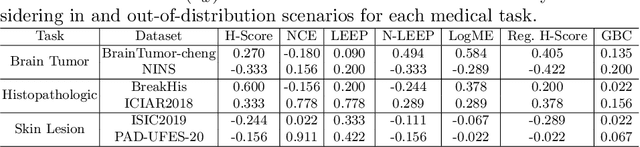
Abstract:Transfer learning boosts the performance of medical image analysis by enabling deep learning (DL) on small datasets through the knowledge acquired from large ones. As the number of DL architectures explodes, exhaustively attempting all candidates becomes unfeasible, motivating cheaper alternatives for choosing them. Transferability scoring methods emerge as an enticing solution, allowing to efficiently calculate a score that correlates with the architecture accuracy on any target dataset. However, since transferability scores have not been evaluated on medical datasets, their use in this context remains uncertain, preventing them from benefiting practitioners. We fill that gap in this work, thoroughly evaluating seven transferability scores in three medical applications, including out-of-distribution scenarios. Despite promising results in general-purpose datasets, our results show that no transferability score can reliably and consistently estimate target performance in medical contexts, inviting further work in that direction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge