Ghassan Hamarneh
IMA++: ISIC Archive Multi-Annotator Dermoscopic Skin Lesion Segmentation Dataset
Dec 25, 2025Abstract:Multi-annotator medical image segmentation is an important research problem, but requires annotated datasets that are expensive to collect. Dermoscopic skin lesion imaging allows human experts and AI systems to observe morphological structures otherwise not discernable from regular clinical photographs. However, currently there are no large-scale publicly available multi-annotator skin lesion segmentation (SLS) datasets with annotator-labels for dermoscopic skin lesion imaging. We introduce ISIC MultiAnnot++, a large public multi-annotator skin lesion segmentation dataset for images from the ISIC Archive. The final dataset contains 17,684 segmentation masks spanning 14,967 dermoscopic images, where 2,394 dermoscopic images have 2-5 segmentations per image, making it the largest publicly available SLS dataset. Further, metadata about the segmentation, including the annotators' skill level and segmentation tool, is included, enabling research on topics such as annotator-specific preference modeling for segmentation and annotator metadata analysis. We provide an analysis on the characteristics of this dataset, curated data partitions, and consensus segmentation masks.
Learning What NOT to Count
Apr 16, 2025Abstract:Few/zero-shot object counting methods reduce the need for extensive annotations but often struggle to distinguish between fine-grained categories, especially when multiple similar objects appear in the same scene. To address this limitation, we propose an annotation-free approach that enables the seamless integration of new fine-grained categories into existing few/zero-shot counting models. By leveraging latent generative models, we synthesize high-quality, category-specific crowded scenes, providing a rich training source for adapting to new categories without manual labeling. Our approach introduces an attention prediction network that identifies fine-grained category boundaries trained using only synthetic pseudo-annotated data. At inference, these fine-grained attention estimates refine the output of existing few/zero-shot counting networks. To benchmark our method, we further introduce the FGTC dataset, a taxonomy-specific fine-grained object counting dataset for natural images. Our method substantially enhances pre-trained state-of-the-art models on fine-grained taxon counting tasks, while using only synthetic data. Code and data to be released upon acceptance.
Disentangled PET Lesion Segmentation
Nov 04, 2024


Abstract:PET imaging is an invaluable tool in clinical settings as it captures the functional activity of both healthy anatomy and cancerous lesions. Developing automatic lesion segmentation methods for PET images is crucial since manual lesion segmentation is laborious and prone to inter- and intra-observer variability. We propose PET-Disentangler, a 3D disentanglement method that uses a 3D UNet-like encoder-decoder architecture to disentangle disease and normal healthy anatomical features with losses for segmentation, reconstruction, and healthy component plausibility. A critic network is used to encourage the healthy latent features to match the distribution of healthy samples and thus encourages these features to not contain any lesion-related features. Our quantitative results show that PET-Disentangler is less prone to incorrectly declaring healthy and high tracer uptake regions as cancerous lesions, since such uptake pattern would be assigned to the disentangled healthy component.
Debiasify: Self-Distillation for Unsupervised Bias Mitigation
Nov 01, 2024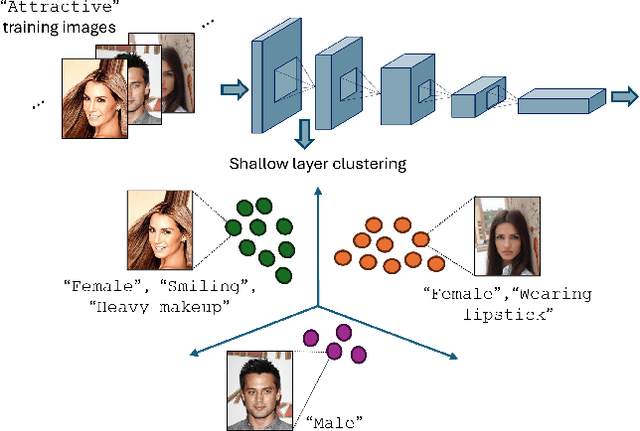
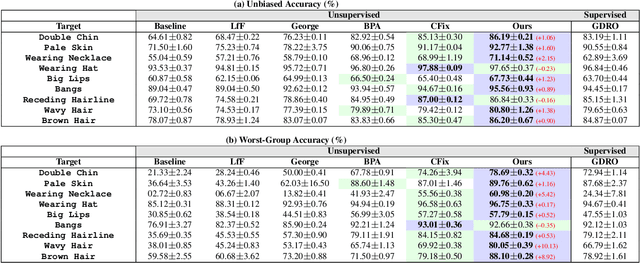
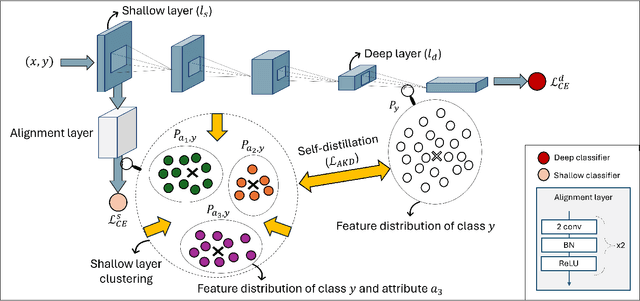
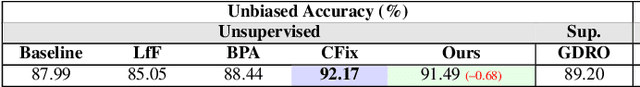
Abstract:Simplicity bias poses a significant challenge in neural networks, often leading models to favor simpler solutions and inadvertently learn decision rules influenced by spurious correlations. This results in biased models with diminished generalizability. While many current approaches depend on human supervision, obtaining annotations for various bias attributes is often impractical. To address this, we introduce Debiasify, a novel self-distillation approach that requires no prior knowledge about the nature of biases. Our method leverages a new distillation loss to transfer knowledge within the network, from deeper layers containing complex, highly-predictive features to shallower layers with simpler, attribute-conditioned features in an unsupervised manner. This enables Debiasify to learn robust, debiased representations that generalize effectively across diverse biases and datasets, improving both worst-group performance and overall accuracy. Extensive experiments on computer vision and medical imaging benchmarks demonstrate the effectiveness of our approach, significantly outperforming previous unsupervised debiasing methods (e.g., a 10.13% improvement in worst-group accuracy for Wavy Hair classification in CelebA) and achieving comparable or superior performance to supervised approaches. Our code is publicly available at the following link: Debiasify.
SMITE: Segment Me In TimE
Oct 24, 2024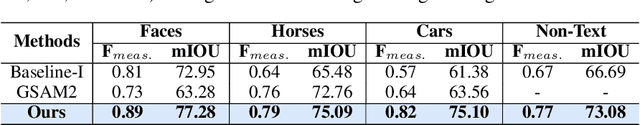
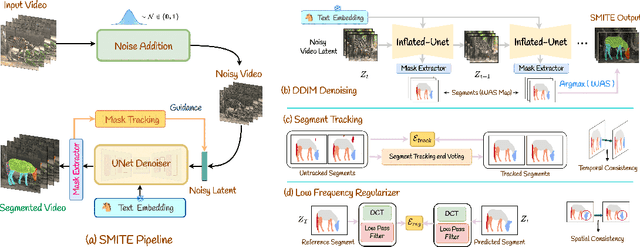
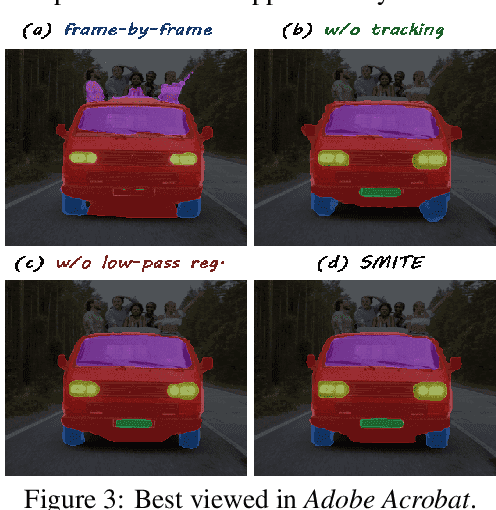
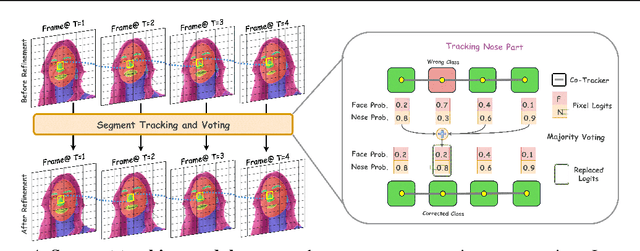
Abstract:Segmenting an object in a video presents significant challenges. Each pixel must be accurately labelled, and these labels must remain consistent across frames. The difficulty increases when the segmentation is with arbitrary granularity, meaning the number of segments can vary arbitrarily, and masks are defined based on only one or a few sample images. In this paper, we address this issue by employing a pre-trained text to image diffusion model supplemented with an additional tracking mechanism. We demonstrate that our approach can effectively manage various segmentation scenarios and outperforms state-of-the-art alternatives.
Segmentation Style Discovery: Application to Skin Lesion Images
Aug 05, 2024Abstract:Variability in medical image segmentation, arising from annotator preferences, expertise, and their choice of tools, has been well documented. While the majority of multi-annotator segmentation approaches focus on modeling annotator-specific preferences, they require annotator-segmentation correspondence. In this work, we introduce the problem of segmentation style discovery, and propose StyleSeg, a segmentation method that learns plausible, diverse, and semantically consistent segmentation styles from a corpus of image-mask pairs without any knowledge of annotator correspondence. StyleSeg consistently outperforms competing methods on four publicly available skin lesion segmentation (SLS) datasets. We also curate ISIC-MultiAnnot, the largest multi-annotator SLS dataset with annotator correspondence, and our results show a strong alignment, using our newly proposed measure AS2, between the predicted styles and annotator preferences. The code and the dataset are available at https://github.com/sfu-mial/StyleSeg.
Lesion Elevation Prediction from Skin Images Improves Diagnosis
Aug 05, 2024Abstract:While deep learning-based computer-aided diagnosis for skin lesion image analysis is approaching dermatologists' performance levels, there are several works showing that incorporating additional features such as shape priors, texture, color constancy, and illumination further improves the lesion diagnosis performance. In this work, we look at another clinically useful feature, skin lesion elevation, and investigate the feasibility of predicting and leveraging skin lesion elevation labels. Specifically, we use a deep learning model to predict image-level lesion elevation labels from 2D skin lesion images. We test the elevation prediction accuracy on the derm7pt dataset, and use the elevation prediction model to estimate elevation labels for images from five other datasets: ISIC 2016, 2017, and 2018 Challenge datasets, MSK, and DermoFit. We evaluate cross-domain generalization by using these estimated elevation labels as auxiliary inputs to diagnosis models, and show that these improve the classification performance, with AUROC improvements of up to 6.29% and 2.69% for dermoscopic and clinical images, respectively. The code is publicly available at https://github.com/sfu-mial/LesionElevation.
BiasPruner: Debiased Continual Learning for Medical Image Classification
Jul 11, 2024Abstract:Continual Learning (CL) is crucial for enabling networks to dynamically adapt as they learn new tasks sequentially, accommodating new data and classes without catastrophic forgetting. Diverging from conventional perspectives on CL, our paper introduces a new perspective wherein forgetting could actually benefit the sequential learning paradigm. Specifically, we present BiasPruner, a CL framework that intentionally forgets spurious correlations in the training data that could lead to shortcut learning. Utilizing a new bias score that measures the contribution of each unit in the network to learning spurious features, BiasPruner prunes those units with the highest bias scores to form a debiased subnetwork preserved for a given task. As BiasPruner learns a new task, it constructs a new debiased subnetwork, potentially incorporating units from previous subnetworks, which improves adaptation and performance on the new task. During inference, BiasPruner employs a simple task-agnostic approach to select the best debiased subnetwork for predictions. We conduct experiments on three medical datasets for skin lesion classification and chest X-Ray classification and demonstrate that BiasPruner consistently outperforms SOTA CL methods in terms of classification performance and fairness. Our code is available here.
Representing Anatomical Trees by Denoising Diffusion of Implicit Neural Fields
Mar 13, 2024Abstract:Anatomical trees play a central role in clinical diagnosis and treatment planning. However, accurately representing anatomical trees is challenging due to their varying and complex topology and geometry. Traditional methods for representing tree structures, captured using medical imaging, while invaluable for visualizing vascular and bronchial networks, exhibit drawbacks in terms of limited resolution, flexibility, and efficiency. Recently, implicit neural representations (INRs) have emerged as a powerful tool for representing shapes accurately and efficiently. We propose a novel approach for representing anatomical trees using INR, while also capturing the distribution of a set of trees via denoising diffusion in the space of INRs. We accurately capture the intricate geometries and topologies of anatomical trees at any desired resolution. Through extensive qualitative and quantitative evaluation, we demonstrate high-fidelity tree reconstruction with arbitrary resolution yet compact storage, and versatility across anatomical sites and tree complexities.
AFreeCA: Annotation-Free Counting for All
Mar 07, 2024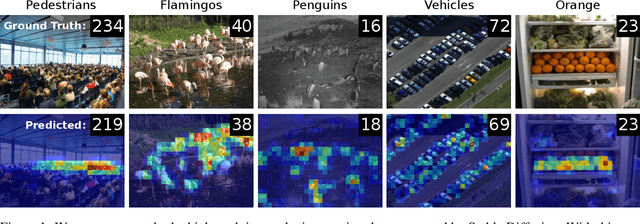
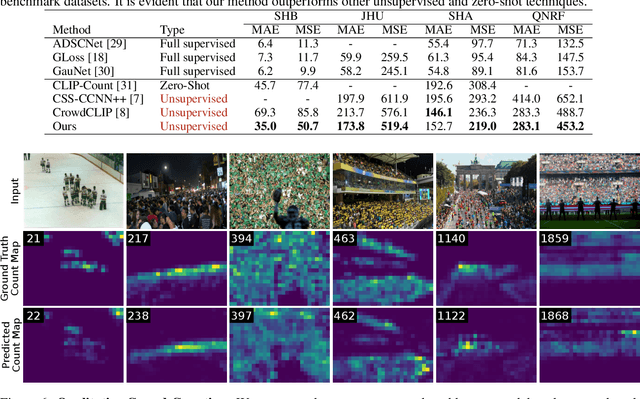
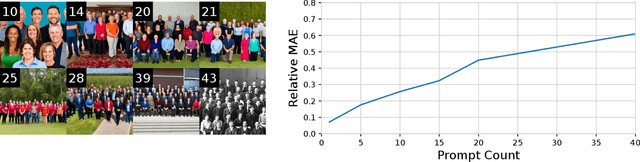
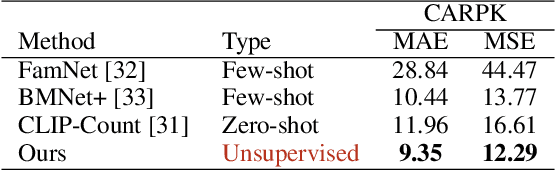
Abstract:Object counting methods typically rely on manually annotated datasets. The cost of creating such datasets has restricted the versatility of these networks to count objects from specific classes (such as humans or penguins), and counting objects from diverse categories remains a challenge. The availability of robust text-to-image latent diffusion models (LDMs) raises the question of whether these models can be utilized to generate counting datasets. However, LDMs struggle to create images with an exact number of objects based solely on text prompts but they can be used to offer a dependable \textit{sorting} signal by adding and removing objects within an image. Leveraging this data, we initially introduce an unsupervised sorting methodology to learn object-related features that are subsequently refined and anchored for counting purposes using counting data generated by LDMs. Further, we present a density classifier-guided method for dividing an image into patches containing objects that can be reliably counted. Consequently, we can generate counting data for any type of object and count them in an unsupervised manner. Our approach outperforms other unsupervised and few-shot alternatives and is not restricted to specific object classes for which counting data is available. Code to be released upon acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge