Hanene Ben Yedder
Disentangled PET Lesion Segmentation
Nov 04, 2024


Abstract:PET imaging is an invaluable tool in clinical settings as it captures the functional activity of both healthy anatomy and cancerous lesions. Developing automatic lesion segmentation methods for PET images is crucial since manual lesion segmentation is laborious and prone to inter- and intra-observer variability. We propose PET-Disentangler, a 3D disentanglement method that uses a 3D UNet-like encoder-decoder architecture to disentangle disease and normal healthy anatomical features with losses for segmentation, reconstruction, and healthy component plausibility. A critic network is used to encourage the healthy latent features to match the distribution of healthy samples and thus encourages these features to not contain any lesion-related features. Our quantitative results show that PET-Disentangler is less prone to incorrectly declaring healthy and high tracer uptake regions as cancerous lesions, since such uptake pattern would be assigned to the disentangled healthy component.
Benchmarking Transcriptomics Foundation Models for Perturbation Analysis : one PCA still rules them all
Oct 17, 2024
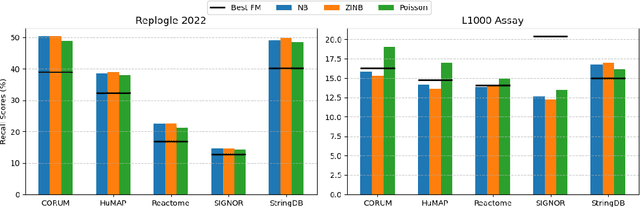
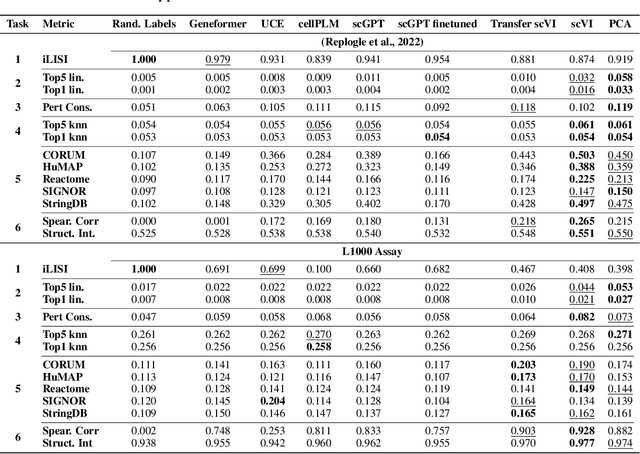
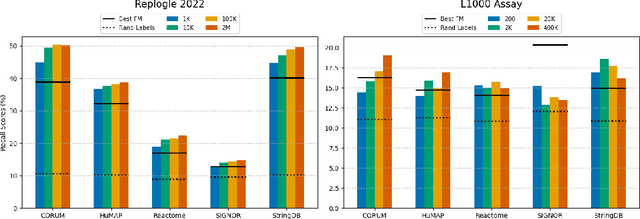
Abstract:Understanding the relationships among genes, compounds, and their interactions in living organisms remains limited due to technological constraints and the complexity of biological data. Deep learning has shown promise in exploring these relationships using various data types. However, transcriptomics, which provides detailed insights into cellular states, is still underused due to its high noise levels and limited data availability. Recent advancements in transcriptomics sequencing provide new opportunities to uncover valuable insights, especially with the rise of many new foundation models for transcriptomics, yet no benchmark has been made to robustly evaluate the effectiveness of these rising models for perturbation analysis. This article presents a novel biologically motivated evaluation framework and a hierarchy of perturbation analysis tasks for comparing the performance of pretrained foundation models to each other and to more classical techniques of learning from transcriptomics data. We compile diverse public datasets from different sequencing techniques and cell lines to assess models performance. Our approach identifies scVI and PCA to be far better suited models for understanding biological perturbations in comparison to existing foundation models, especially in their application in real-world scenarios.
Deep Learning for Biomedical Image Reconstruction: A Survey
Feb 26, 2020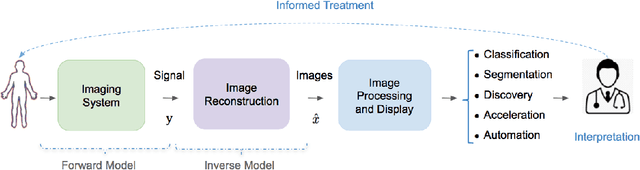
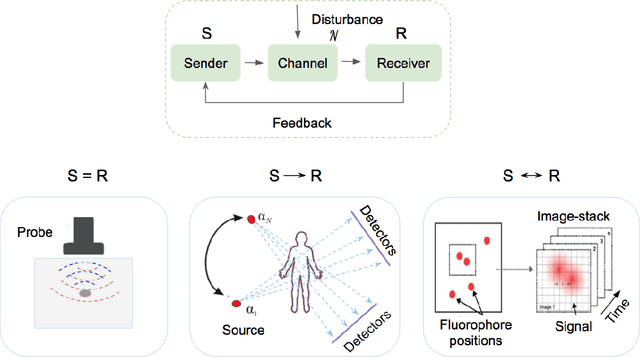
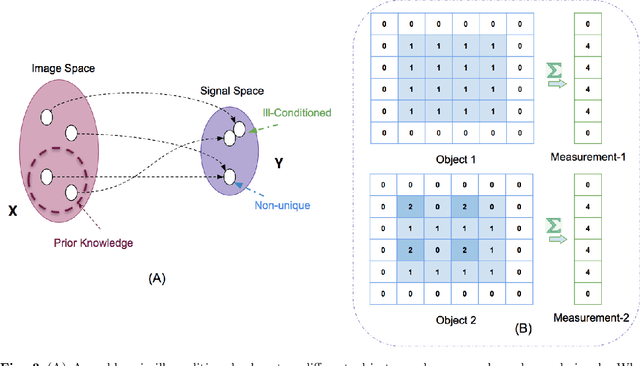
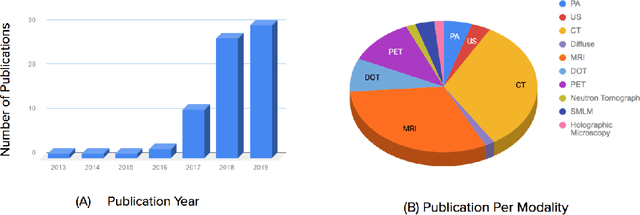
Abstract:Medical imaging is an invaluable resource in medicine as it enables to peer inside the human body and provides scientists and physicians with a wealth of information indispensable for understanding, modelling, diagnosis, and treatment of diseases. Reconstruction algorithms entail transforming signals collected by acquisition hardware into interpretable images. Reconstruction is a challenging task given the ill-posed of the problem and the absence of exact analytic inverse transforms in practical cases. While the last decades witnessed impressive advancements in terms of new modalities, improved temporal and spatial resolution, reduced cost, and wider applicability, several improvements can still be envisioned such as reducing acquisition and reconstruction time to reduce patient's exposure to radiation and discomfort while increasing clinics throughput and reconstruction accuracy. Furthermore, the deployment of biomedical imaging in handheld devices with small power requires a fine balance between accuracy and latency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge