Kumar Abhishek
IMA++: ISIC Archive Multi-Annotator Dermoscopic Skin Lesion Segmentation Dataset
Dec 25, 2025Abstract:Multi-annotator medical image segmentation is an important research problem, but requires annotated datasets that are expensive to collect. Dermoscopic skin lesion imaging allows human experts and AI systems to observe morphological structures otherwise not discernable from regular clinical photographs. However, currently there are no large-scale publicly available multi-annotator skin lesion segmentation (SLS) datasets with annotator-labels for dermoscopic skin lesion imaging. We introduce ISIC MultiAnnot++, a large public multi-annotator skin lesion segmentation dataset for images from the ISIC Archive. The final dataset contains 17,684 segmentation masks spanning 14,967 dermoscopic images, where 2,394 dermoscopic images have 2-5 segmentations per image, making it the largest publicly available SLS dataset. Further, metadata about the segmentation, including the annotators' skill level and segmentation tool, is included, enabling research on topics such as annotator-specific preference modeling for segmentation and annotator metadata analysis. We provide an analysis on the characteristics of this dataset, curated data partitions, and consensus segmentation masks.
Disentangled PET Lesion Segmentation
Nov 04, 2024


Abstract:PET imaging is an invaluable tool in clinical settings as it captures the functional activity of both healthy anatomy and cancerous lesions. Developing automatic lesion segmentation methods for PET images is crucial since manual lesion segmentation is laborious and prone to inter- and intra-observer variability. We propose PET-Disentangler, a 3D disentanglement method that uses a 3D UNet-like encoder-decoder architecture to disentangle disease and normal healthy anatomical features with losses for segmentation, reconstruction, and healthy component plausibility. A critic network is used to encourage the healthy latent features to match the distribution of healthy samples and thus encourages these features to not contain any lesion-related features. Our quantitative results show that PET-Disentangler is less prone to incorrectly declaring healthy and high tracer uptake regions as cancerous lesions, since such uptake pattern would be assigned to the disentangled healthy component.
Lesion Elevation Prediction from Skin Images Improves Diagnosis
Aug 05, 2024Abstract:While deep learning-based computer-aided diagnosis for skin lesion image analysis is approaching dermatologists' performance levels, there are several works showing that incorporating additional features such as shape priors, texture, color constancy, and illumination further improves the lesion diagnosis performance. In this work, we look at another clinically useful feature, skin lesion elevation, and investigate the feasibility of predicting and leveraging skin lesion elevation labels. Specifically, we use a deep learning model to predict image-level lesion elevation labels from 2D skin lesion images. We test the elevation prediction accuracy on the derm7pt dataset, and use the elevation prediction model to estimate elevation labels for images from five other datasets: ISIC 2016, 2017, and 2018 Challenge datasets, MSK, and DermoFit. We evaluate cross-domain generalization by using these estimated elevation labels as auxiliary inputs to diagnosis models, and show that these improve the classification performance, with AUROC improvements of up to 6.29% and 2.69% for dermoscopic and clinical images, respectively. The code is publicly available at https://github.com/sfu-mial/LesionElevation.
Segmentation Style Discovery: Application to Skin Lesion Images
Aug 05, 2024Abstract:Variability in medical image segmentation, arising from annotator preferences, expertise, and their choice of tools, has been well documented. While the majority of multi-annotator segmentation approaches focus on modeling annotator-specific preferences, they require annotator-segmentation correspondence. In this work, we introduce the problem of segmentation style discovery, and propose StyleSeg, a segmentation method that learns plausible, diverse, and semantically consistent segmentation styles from a corpus of image-mask pairs without any knowledge of annotator correspondence. StyleSeg consistently outperforms competing methods on four publicly available skin lesion segmentation (SLS) datasets. We also curate ISIC-MultiAnnot, the largest multi-annotator SLS dataset with annotator correspondence, and our results show a strong alignment, using our newly proposed measure AS2, between the predicted styles and annotator preferences. The code and the dataset are available at https://github.com/sfu-mial/StyleSeg.
Investigating the Quality of DermaMNIST and Fitzpatrick17k Dermatological Image Datasets
Jan 25, 2024Abstract:The remarkable progress of deep learning in dermatological tasks has brought us closer to achieving diagnostic accuracies comparable to those of human experts. However, while large datasets play a crucial role in the development of reliable deep neural network models, the quality of data therein and their correct usage are of paramount importance. Several factors can impact data quality, such as the presence of duplicates, data leakage across train-test partitions, mislabeled images, and the absence of a well-defined test partition. In this paper, we conduct meticulous analyses of two popular dermatological image datasets: DermaMNIST and Fitzpatrick17k, uncovering these data quality issues, measure the effects of these problems on the benchmark results, and propose corrections to the datasets. Besides ensuring the reproducibility of our analysis, by making our analysis pipeline and the accompanying code publicly available, we aim to encourage similar explorations and to facilitate the identification and addressing of potential data quality issues in other large datasets.
DermSynth3D: Synthesis of in-the-wild Annotated Dermatology Images
May 25, 2023



Abstract:In recent years, deep learning (DL) has shown great potential in the field of dermatological image analysis. However, existing datasets in this domain have significant limitations, including a small number of image samples, limited disease conditions, insufficient annotations, and non-standardized image acquisitions. To address these shortcomings, we propose a novel framework called DermSynth3D. DermSynth3D blends skin disease patterns onto 3D textured meshes of human subjects using a differentiable renderer and generates 2D images from various camera viewpoints under chosen lighting conditions in diverse background scenes. Our method adheres to top-down rules that constrain the blending and rendering process to create 2D images with skin conditions that mimic in-the-wild acquisitions, ensuring more meaningful results. The framework generates photo-realistic 2D dermoscopy images and the corresponding dense annotations for semantic segmentation of the skin, skin conditions, body parts, bounding boxes around lesions, depth maps, and other 3D scene parameters, such as camera position and lighting conditions. DermSynth3D allows for the creation of custom datasets for various dermatology tasks. We demonstrate the effectiveness of data generated using DermSynth3D by training DL models on synthetic data and evaluating them on various dermatology tasks using real 2D dermatological images. We make our code publicly available at https://github.com/sfu-mial/DermSynth3D.
Attribution-based XAI Methods in Computer Vision: A Review
Nov 27, 2022Abstract:The advancements in deep learning-based methods for visual perception tasks have seen astounding growth in the last decade, with widespread adoption in a plethora of application areas from autonomous driving to clinical decision support systems. Despite their impressive performance, these deep learning-based models remain fairly opaque in their decision-making process, making their deployment in human-critical tasks a risky endeavor. This in turn makes understanding the decisions made by these models crucial for their reliable deployment. Explainable AI (XAI) methods attempt to address this by offering explanations for such black-box deep learning methods. In this paper, we provide a comprehensive survey of attribution-based XAI methods in computer vision and review the existing literature for gradient-based, perturbation-based, and contrastive methods for XAI, and provide insights on the key challenges in developing and evaluating robust XAI methods.
CIRCLe: Color Invariant Representation Learning for Unbiased Classification of Skin Lesions
Aug 29, 2022

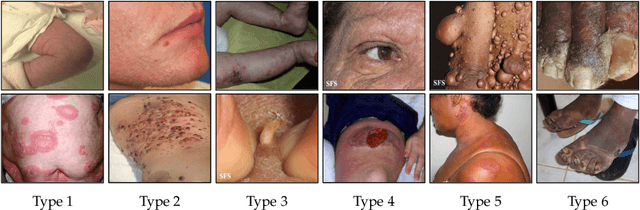
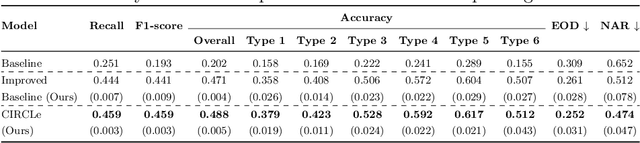
Abstract:While deep learning based approaches have demonstrated expert-level performance in dermatological diagnosis tasks, they have also been shown to exhibit biases toward certain demographic attributes, particularly skin types (e.g., light versus dark), a fairness concern that must be addressed. We propose CIRCLe, a skin color invariant deep representation learning method for improving fairness in skin lesion classification. CIRCLe is trained to classify images by utilizing a regularization loss that encourages images with the same diagnosis but different skin types to have similar latent representations. Through extensive evaluation and ablation studies, we demonstrate CIRCLe's superior performance over the state-of-the-art when evaluated on 16k+ images spanning 6 Fitzpatrick skin types and 114 diseases, using classification accuracy, equal opportunity difference (for light versus dark groups), and normalized accuracy range, a new measure we propose to assess fairness on multiple skin type groups.
A Survey on Deep Learning for Skin Lesion Segmentation
Jun 01, 2022



Abstract:Skin cancer is a major public health problem that could benefit from computer-aided diagnosis to reduce the burden of this common disease. Skin lesion segmentation from images is an important step toward achieving this goal. However, the presence of natural and artificial artifacts (e.g., hair and air bubbles), intrinsic factors (e.g., lesion shape and contrast), and variations in image acquisition conditions make skin lesion segmentation a challenging task. Recently, various researchers have explored the applicability of deep learning models to skin lesion segmentation. In this survey, we cross-examine 134 research papers that deal with deep learning based segmentation of skin lesions. We analyze these works along several dimensions, including input data (datasets, preprocessing, and synthetic data generation), model design (architecture, modules, and losses), and evaluation aspects (data annotation requirements and segmentation performance). We discuss these dimensions both from the viewpoint of select seminal works, and from a systematic viewpoint, examining how those choices have influenced current trends, and how their limitations should be addressed. We summarize all examined works in a comprehensive table to facilitate comparisons.
Multi-Sample $ζ$-mixup: Richer, More Realistic Synthetic Samples from a $p$-Series Interpolant
Apr 07, 2022



Abstract:Modern deep learning training procedures rely on model regularization techniques such as data augmentation methods, which generate training samples that increase the diversity of data and richness of label information. A popular recent method, mixup, uses convex combinations of pairs of original samples to generate new samples. However, as we show in our experiments, mixup can produce undesirable synthetic samples, where the data is sampled off the manifold and can contain incorrect labels. We propose $\zeta$-mixup, a generalization of mixup with provably and demonstrably desirable properties that allows convex combinations of $N \geq 2$ samples, leading to more realistic and diverse outputs that incorporate information from $N$ original samples by using a $p$-series interpolant. We show that, compared to mixup, $\zeta$-mixup better preserves the intrinsic dimensionality of the original datasets, which is a desirable property for training generalizable models. Furthermore, we show that our implementation of $\zeta$-mixup is faster than mixup, and extensive evaluation on controlled synthetic and 24 real-world natural and medical image classification datasets shows that $\zeta$-mixup outperforms mixup and traditional data augmentation techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge