Zahra Mirikharaji
A Survey on Deep Learning for Skin Lesion Segmentation
Jun 01, 2022



Abstract:Skin cancer is a major public health problem that could benefit from computer-aided diagnosis to reduce the burden of this common disease. Skin lesion segmentation from images is an important step toward achieving this goal. However, the presence of natural and artificial artifacts (e.g., hair and air bubbles), intrinsic factors (e.g., lesion shape and contrast), and variations in image acquisition conditions make skin lesion segmentation a challenging task. Recently, various researchers have explored the applicability of deep learning models to skin lesion segmentation. In this survey, we cross-examine 134 research papers that deal with deep learning based segmentation of skin lesions. We analyze these works along several dimensions, including input data (datasets, preprocessing, and synthetic data generation), model design (architecture, modules, and losses), and evaluation aspects (data annotation requirements and segmentation performance). We discuss these dimensions both from the viewpoint of select seminal works, and from a systematic viewpoint, examining how those choices have influenced current trends, and how their limitations should be addressed. We summarize all examined works in a comprehensive table to facilitate comparisons.
D-LEMA: Deep Learning Ensembles from Multiple Annotations -- Application to Skin Lesion Segmentation
Dec 14, 2020



Abstract:Medical image segmentation annotations suffer from inter/intra-observer variations even among experts due to intrinsic differences in human annotators and ambiguous boundaries. Leveraging a collection of annotators' opinions for an image is an interesting way of estimating a gold standard. Although training deep models in a supervised setting with a single annotation per image has been extensively studied, generalizing their training to work with data sets containing multiple annotations per image remains a fairly unexplored problem. In this paper, we propose an approach to handle annotators' disagreements when training a deep model. To this end, we propose an ensemble of Bayesian fully convolutional networks (FCNs) for the segmentation task by considering two major factors in the aggregation of multiple ground truth annotations: (1) handling contradictory annotations in the training data originating from inter-annotator disagreements and (2) improving confidence calibration through the fusion of base models predictions. We demonstrate the superior performance of our approach on the ISIC Archive and explore the generalization performance of our proposed method by cross-data set evaluation on the PH2 and DermoFit data sets.
WhiteNNer-Blind Image Denoising via Noise Whiteness Priors
Aug 08, 2019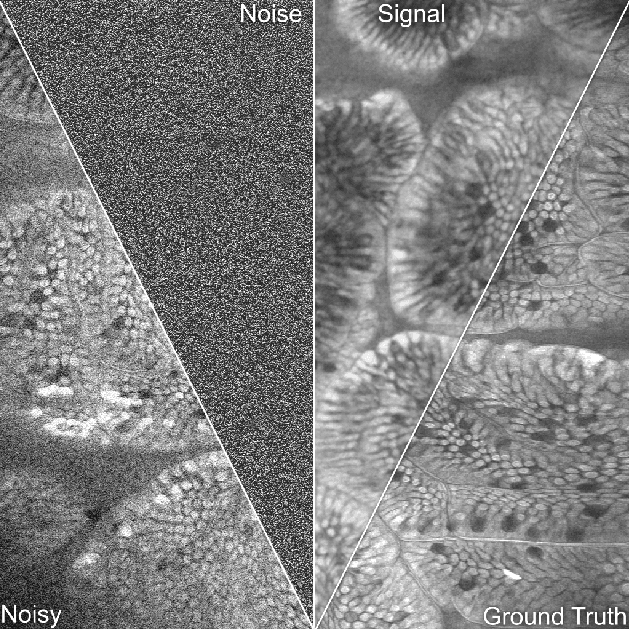
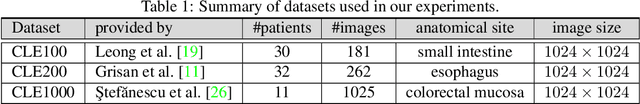
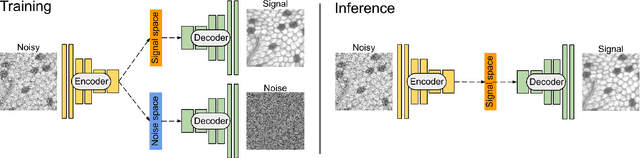
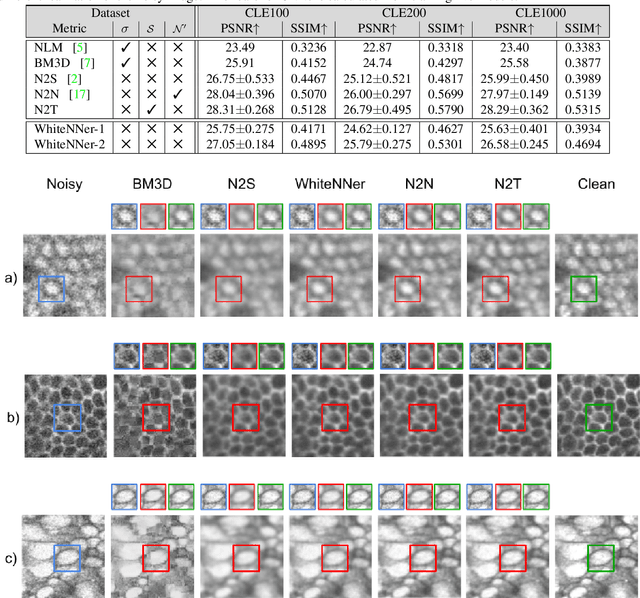
Abstract:The accuracy of medical imaging-based diagnostics is directly impacted by the quality of the collected images. A passive approach to improve image quality is one that lags behind improvements in imaging hardware, awaiting better sensor technology of acquisition devices. An alternative, active strategy is to utilize prior knowledge of the imaging system to directly post-process and improve the acquired images. Traditionally, priors about the image properties are taken into account to restrict the solution space. However, few techniques exploit the prior about the noise properties. In this paper, we propose a neural network-based model for disentangling the signal and noise components of an input noisy image, without the need for any ground truth training data. We design a unified loss function that encodes priors about signal as well as noise estimate in the form of regularization terms. Specifically, by using total variation and piecewise constancy priors along with noise whiteness priors such as auto-correlation and stationary losses, our network learns to decouple an input noisy image into the underlying signal and noise components. We compare our proposed method to Noise2Noise and Noise2Self, as well as non-local mean and BM3D, on three public confocal laser endomicroscopy datasets. Experimental results demonstrate the superiority of our network compared to state-of-the-art in terms of PSNR and SSIM.
Learning to Segment Skin Lesions from Noisy Annotations
Jun 10, 2019


Abstract:Deep convolutional neural networks have driven substantial advancements in the automatic understanding of images. Requiring a large collection of images and their associated annotations is one of the main bottlenecks limiting the adoption of deep networks. In the task of medical image segmentation, requiring pixel level semantic annotations performed by human experts exacerbate this difficulty. This paper proposes a new framework to train a fully convolutional segmentation network from a large set of cheap unreliable annotations and a small set of expert level clean annotations. We propose a spatially adaptive reweighting approach to treat clean and noisy pixel-level annotations commensurately in the loss function. We deploy a meta-learning approach to assign higher importance to pixels whose loss gradient direction is closer to those of clean data. Our experiments on training the network using segmentation ground truth corrupted with different levels of annotation noise show how spatial reweighting improves the robustness of deep networks to noisy annotations.
Star Shape Prior in Fully Convolutional Networks for Skin Lesion Segmentation
Jun 21, 2018


Abstract:Semantic segmentation is an important preliminary step towards automatic medical image interpretation. Recently deep convolutional neural networks have become the first choice for the task of pixel-wise class prediction. While incorporating prior knowledge about the structure of target objects has proven effective in traditional energy-based segmentation approaches, there has not been a clear way for encoding prior knowledge into deep learning frameworks. In this work, we propose a new loss term that encodes the star shape prior into the loss function of an end-to-end trainable fully convolutional network (FCN) framework. We penalize non-star shape segments in FCN prediction maps to guarantee a global structure in segmentation results. Our experiments demonstrate the advantage of regularizing FCN parameters by the star shape prior and our results on the ISBI 2017 skin segmentation challenge data set achieve the first rank in the segmentation task among $21$ participating teams.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge