Saeed Izadi
D-LEMA: Deep Learning Ensembles from Multiple Annotations -- Application to Skin Lesion Segmentation
Dec 14, 2020



Abstract:Medical image segmentation annotations suffer from inter/intra-observer variations even among experts due to intrinsic differences in human annotators and ambiguous boundaries. Leveraging a collection of annotators' opinions for an image is an interesting way of estimating a gold standard. Although training deep models in a supervised setting with a single annotation per image has been extensively studied, generalizing their training to work with data sets containing multiple annotations per image remains a fairly unexplored problem. In this paper, we propose an approach to handle annotators' disagreements when training a deep model. To this end, we propose an ensemble of Bayesian fully convolutional networks (FCNs) for the segmentation task by considering two major factors in the aggregation of multiple ground truth annotations: (1) handling contradictory annotations in the training data originating from inter-annotator disagreements and (2) improving confidence calibration through the fusion of base models predictions. We demonstrate the superior performance of our approach on the ISIC Archive and explore the generalization performance of our proposed method by cross-data set evaluation on the PH2 and DermoFit data sets.
Patch-based Non-Local Bayesian Networks for Blind Confocal Microscopy Denoising
Mar 25, 2020



Abstract:Confocal microscopy is essential for histopathologic cell visualization and quantification. Despite its significant role in biology, fluorescence confocal microscopy suffers from the presence of inherent noise during image acquisition. Non-local patch-wise Bayesian mean filtering (NLB) was until recently the state-of-the-art denoising approach. However, classic denoising methods have been outperformed by neural networks in recent years. In this work, we propose to exploit the strengths of NLB in the framework of Bayesian deep learning. We do so by designing a convolutional neural network and training it to learn parameters of a Gaussian model approximating the prior on noise-free patches given their nearest, similar yet non-local, neighbors. We then apply Bayesian reasoning to leverage the prior and information from the noisy patch in the process of approximating the noise-free patch. Specifically, we use the closed-form analytic \textit{maximum a posteriori} (MAP) estimate in the NLB algorithm to obtain the noise-free patch that maximizes the posterior distribution. The performance of our proposed method is evaluated on confocal microscopy images with real noise Poisson-Gaussian noise. Our experiments reveal the superiority of our approach against state-of-the-art unsupervised denoising techniques.
WhiteNNer-Blind Image Denoising via Noise Whiteness Priors
Aug 08, 2019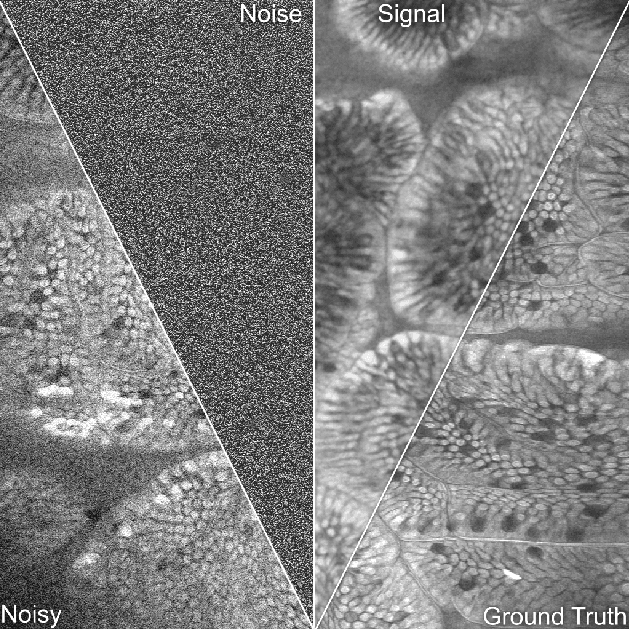
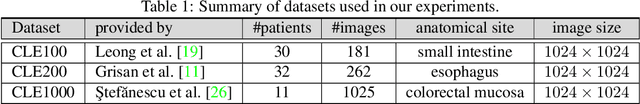
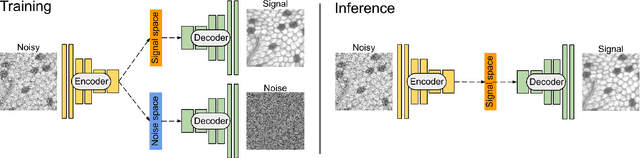
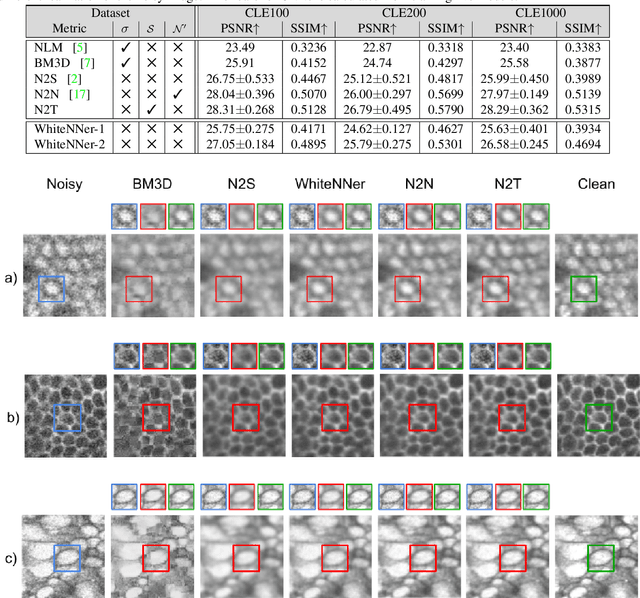
Abstract:The accuracy of medical imaging-based diagnostics is directly impacted by the quality of the collected images. A passive approach to improve image quality is one that lags behind improvements in imaging hardware, awaiting better sensor technology of acquisition devices. An alternative, active strategy is to utilize prior knowledge of the imaging system to directly post-process and improve the acquired images. Traditionally, priors about the image properties are taken into account to restrict the solution space. However, few techniques exploit the prior about the noise properties. In this paper, we propose a neural network-based model for disentangling the signal and noise components of an input noisy image, without the need for any ground truth training data. We design a unified loss function that encodes priors about signal as well as noise estimate in the form of regularization terms. Specifically, by using total variation and piecewise constancy priors along with noise whiteness priors such as auto-correlation and stationary losses, our network learns to decouple an input noisy image into the underlying signal and noise components. We compare our proposed method to Noise2Noise and Noise2Self, as well as non-local mean and BM3D, on three public confocal laser endomicroscopy datasets. Experimental results demonstrate the superiority of our network compared to state-of-the-art in terms of PSNR and SSIM.
Image Super Resolution via Bilinear Pooling: Application to Confocal Endomicroscopy
Jul 23, 2019



Abstract:Recent developments in image acquisition literature have miniaturized the confocal laser endomicroscopes to improve usability and flexibility of the apparatus in actual clinical settings. However, miniaturized devices collect less light and have fewer optical components, resulting in pixelation artifacts and low resolution images. Owing to the strength of deep networks, many supervised methods known as super resolution have achieved considerable success in restoring low resolution images by generating the missing high frequency details. In this work, we propose a novel attention mechanism that, for the first time, combines 1st- and 2nd-order statistics for pooling operation, in the spatial and channel-wise dimensions. We compare the efficacy of our method to 11 other existing single image super resolution techniques that compensate for the reduction in image quality caused by the necessity of endomicroscope miniaturization. All evaluations are carried out on three publicly available datasets. Experimental results show that our method can produce competitive results against state-of-the-art in terms of PSNR, SSIM, and IFC metrics. Additionally, our proposed method contains small number of parameters, which makes it lightweight and fast for real-time applications.
Can Deep Learning Relax Endomicroscopy Hardware Miniaturization Requirements?
Jun 21, 2018



Abstract:Confocal laser endomicroscopy (CLE) is a novel imaging modality that provides in vivo histological cross-sections of examined tissue. Recently, attempts have been made to develop miniaturized in vivo imaging devices, specifically confocal laser microscopes, for both clinical and research applications. However, current implementations of miniature CLE components, such as confocal lenses, compromise image resolution, signal-to-noise ratio, or both, which negatively impacts the utility of in vivo imaging. In this work, we demonstrate that software-based techniques can be used to recover lost information due to endomicroscopy hardware miniaturization and reconstruct images of higher resolution. Particularly, a densely connected convolutional neural network is used to reconstruct a high-resolution CLE image from a low-resolution input. In the proposed network, each layer is directly connected to all subsequent layers, which results in an effective combination of low-level and high-level features and efficient information flow throughout the network. To train and evaluate our network, we use a dataset of 181 high-resolution CLE images. Both quantitative and qualitative results indicate superiority of the proposed network compared to traditional interpolation techniques and competing learning-based methods. This work demonstrates that software-based super-resolution is a viable approach to compensate for loss of resolution due to endoscopic hardware miniaturization.
What can we learn about CNNs from a large scale controlled object dataset?
Jan 26, 2016



Abstract:Tolerance to image variations (e.g. translation, scale, pose, illumination) is an important desired property of any object recognition system, be it human or machine. Moving towards increasingly bigger datasets has been trending in computer vision specially with the emergence of highly popular deep learning models. While being very useful for learning invariance to object inter- and intra-class shape variability, these large-scale wild datasets are not very useful for learning invariance to other parameters forcing researchers to resort to other tricks for training a model. In this work, we introduce a large-scale synthetic dataset, which is freely and publicly available, and use it to answer several fundamental questions regarding invariance and selectivity properties of convolutional neural networks. Our dataset contains two parts: a) objects shot on a turntable: 16 categories, 8 rotation angles, 11 cameras on a semicircular arch, 5 lighting conditions, 3 focus levels, variety of backgrounds (23.4 per instance) generating 1320 images per instance (over 20 million images in total), and b) scenes: in which a robot arm takes pictures of objects on a 1:160 scale scene. We study: 1) invariance and selectivity of different CNN layers, 2) knowledge transfer from one object category to another, 3) systematic or random sampling of images to build a train set, 4) domain adaptation from synthetic to natural scenes, and 5) order of knowledge delivery to CNNs. We also explore how our analyses can lead the field to develop more efficient CNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge