Jeremy Kawahara
IMA++: ISIC Archive Multi-Annotator Dermoscopic Skin Lesion Segmentation Dataset
Dec 25, 2025Abstract:Multi-annotator medical image segmentation is an important research problem, but requires annotated datasets that are expensive to collect. Dermoscopic skin lesion imaging allows human experts and AI systems to observe morphological structures otherwise not discernable from regular clinical photographs. However, currently there are no large-scale publicly available multi-annotator skin lesion segmentation (SLS) datasets with annotator-labels for dermoscopic skin lesion imaging. We introduce ISIC MultiAnnot++, a large public multi-annotator skin lesion segmentation dataset for images from the ISIC Archive. The final dataset contains 17,684 segmentation masks spanning 14,967 dermoscopic images, where 2,394 dermoscopic images have 2-5 segmentations per image, making it the largest publicly available SLS dataset. Further, metadata about the segmentation, including the annotators' skill level and segmentation tool, is included, enabling research on topics such as annotator-specific preference modeling for segmentation and annotator metadata analysis. We provide an analysis on the characteristics of this dataset, curated data partitions, and consensus segmentation masks.
Segmentation Style Discovery: Application to Skin Lesion Images
Aug 05, 2024Abstract:Variability in medical image segmentation, arising from annotator preferences, expertise, and their choice of tools, has been well documented. While the majority of multi-annotator segmentation approaches focus on modeling annotator-specific preferences, they require annotator-segmentation correspondence. In this work, we introduce the problem of segmentation style discovery, and propose StyleSeg, a segmentation method that learns plausible, diverse, and semantically consistent segmentation styles from a corpus of image-mask pairs without any knowledge of annotator correspondence. StyleSeg consistently outperforms competing methods on four publicly available skin lesion segmentation (SLS) datasets. We also curate ISIC-MultiAnnot, the largest multi-annotator SLS dataset with annotator correspondence, and our results show a strong alignment, using our newly proposed measure AS2, between the predicted styles and annotator preferences. The code and the dataset are available at https://github.com/sfu-mial/StyleSeg.
DermSynth3D: Synthesis of in-the-wild Annotated Dermatology Images
May 25, 2023



Abstract:In recent years, deep learning (DL) has shown great potential in the field of dermatological image analysis. However, existing datasets in this domain have significant limitations, including a small number of image samples, limited disease conditions, insufficient annotations, and non-standardized image acquisitions. To address these shortcomings, we propose a novel framework called DermSynth3D. DermSynth3D blends skin disease patterns onto 3D textured meshes of human subjects using a differentiable renderer and generates 2D images from various camera viewpoints under chosen lighting conditions in diverse background scenes. Our method adheres to top-down rules that constrain the blending and rendering process to create 2D images with skin conditions that mimic in-the-wild acquisitions, ensuring more meaningful results. The framework generates photo-realistic 2D dermoscopy images and the corresponding dense annotations for semantic segmentation of the skin, skin conditions, body parts, bounding boxes around lesions, depth maps, and other 3D scene parameters, such as camera position and lighting conditions. DermSynth3D allows for the creation of custom datasets for various dermatology tasks. We demonstrate the effectiveness of data generated using DermSynth3D by training DL models on synthetic data and evaluating them on various dermatology tasks using real 2D dermatological images. We make our code publicly available at https://github.com/sfu-mial/DermSynth3D.
Diffusion-based Data Augmentation for Skin Disease Classification: Impact Across Original Medical Datasets to Fully Synthetic Images
Jan 12, 2023



Abstract:Despite continued advancement in recent years, deep neural networks still rely on large amounts of training data to avoid overfitting. However, labeled training data for real-world applications such as healthcare is limited and difficult to access given longstanding privacy, and strict data sharing policies. By manipulating image datasets in the pixel or feature space, existing data augmentation techniques represent one of the effective ways to improve the quantity and diversity of training data. Here, we look to advance augmentation techniques by building upon the emerging success of text-to-image diffusion probabilistic models in augmenting the training samples of our macroscopic skin disease dataset. We do so by enabling fine-grained control of the image generation process via input text prompts. We demonstrate that this generative data augmentation approach successfully maintains a similar classification accuracy of the visual classifier even when trained on a fully synthetic skin disease dataset. Similar to recent applications of generative models, our study suggests that diffusion models are indeed effective in generating high-quality skin images that do not sacrifice the classifier performance, and can improve the augmentation of training datasets after curation.
Detection and Longitudinal Tracking of Pigmented Skin Lesions in 3D Total-Body Skin Textured Meshes
May 02, 2021
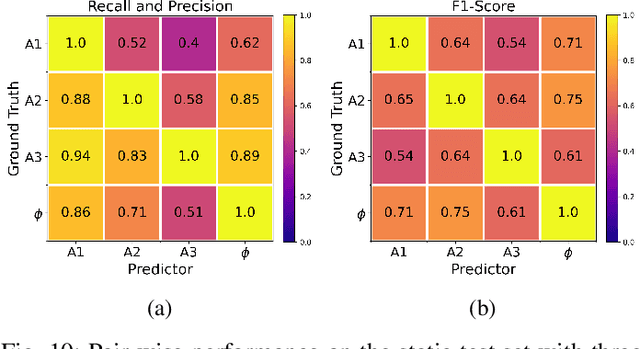
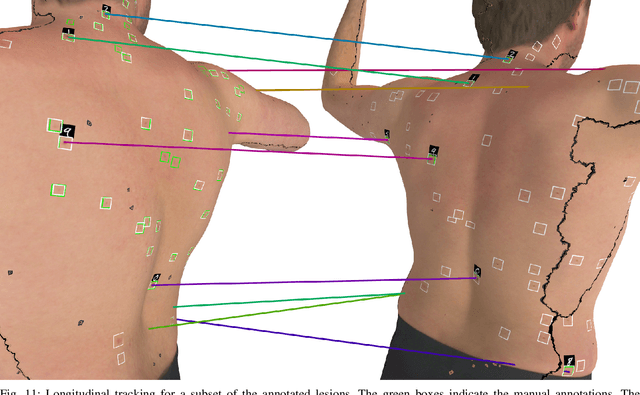

Abstract:We present an automated approach to detect and longitudinally track skin lesions on 3D total-body skin surfaces scans. The acquired 3D mesh of the subject is unwrapped to a 2D texture image, where a trained region convolutional neural network (R-CNN) localizes the lesions within the 2D domain. These detected skin lesions are mapped back to the 3D surface of the subject and, for subjects imaged multiple times, the anatomical correspondences among pairs of meshes and the geodesic distances among lesions are leveraged in our longitudinal lesion tracking algorithm. We evaluated the proposed approach using three sources of data. Firstly, we augmented the 3D meshes of human subjects from the public FAUST dataset with a variety of poses, textures, and images of lesions. Secondly, using a handheld structured light 3D scanner, we imaged a mannequin with multiple synthetic skin lesions at selected location and with varying shapes, sizes, and colours. Finally, we used 3DBodyTex, a publicly available dataset composed of 3D scans imaging the colored (textured) skin of 200 human subjects. We manually annotated locations that appeared to the human eye to contain a pigmented skin lesion as well as tracked a subset of lesions occurring on the same subject imaged in different poses. Our results, on test subjects annotated by three human annotators, suggest that the trained R-CNN detects lesions at a similar performance level as the human annotators. Our lesion tracking algorithm achieves an average accuracy of 80% when identifying corresponding pairs of lesions across subjects imaged in different poses. As there currently is no other large-scale publicly available dataset of 3D total-body skin lesions, we publicly release the 10 mannequin meshes and over 25,000 3DBodyTex manual annotations, which we hope will further research on total-body skin lesion analysis.
Visual Diagnosis of Dermatological Disorders: Human and Machine Performance
Jun 04, 2019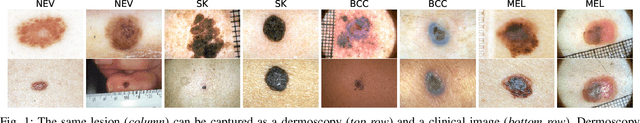
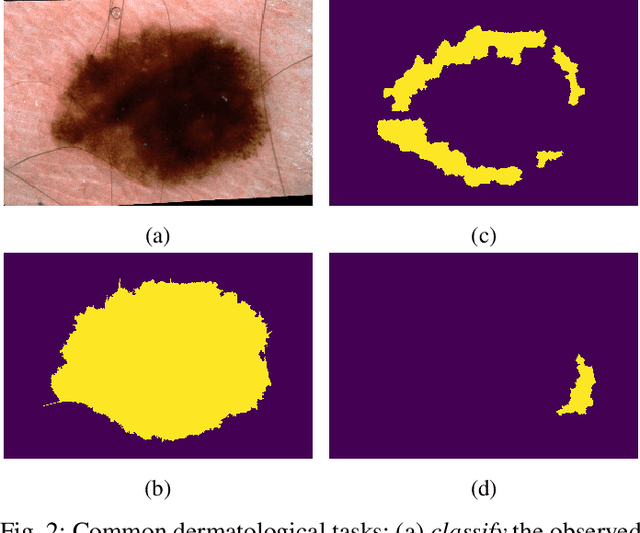
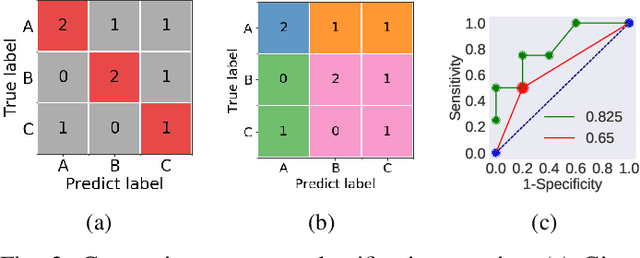
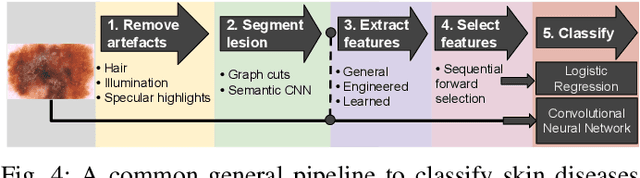
Abstract:Skin conditions are a global health concern, ranking the fourth highest cause of nonfatal disease burden when measured as years lost due to disability. As diagnosing, or classifying, skin diseases can help determine effective treatment, dermatologists have extensively researched how to diagnose conditions from a patient's history and the lesion's visual appearance. Computer vision researchers are attempting to encode this diagnostic ability into machines, and several recent studies report machine level performance comparable with dermatologists. This report reviews machine approaches to classify skin images and consider their performance when compared to human dermatologists. Following an overview of common image modalities, dermatologists' diagnostic approaches and common tasks, and publicly available datasets, we discuss approaches to machine skin lesion classification. We then review works that directly compare human and machine performance. Finally, this report addresses the limitations and sources of errors in image-based skin disease diagnosis, applicable to both machines and dermatologists in a teledermatology setting.
Fully Convolutional Networks to Detect Clinical Dermoscopic Features
Mar 14, 2017


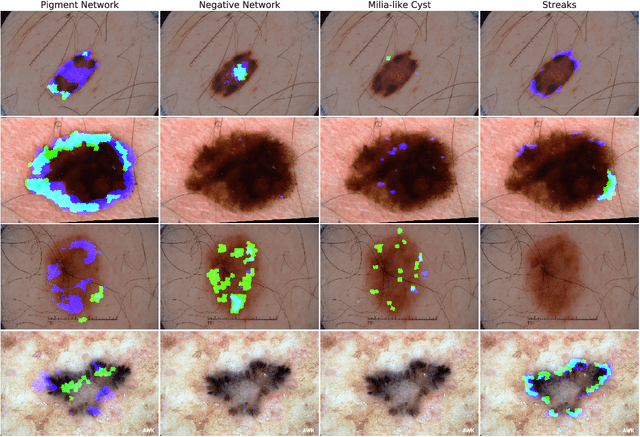
Abstract:We use a pretrained fully convolutional neural network to detect clinical dermoscopic features from dermoscopy skin lesion images. We reformulate the superpixel classification task as an image segmentation problem, and extend a neural network architecture originally designed for image classification to detect dermoscopic features. Specifically, we interpolate the feature maps from several layers in the network to match the size of the input, concatenate the resized feature maps, and train the network to minimize a smoothed negative F1 score. Over the public validation leaderboard of the 2017 ISIC/ISBI Lesion Dermoscopic Feature Extraction Challenge, our approach achieves 89.3% AUROC, the highest averaged score when compared to the other two entries. Results over the private test leaderboard are still to be announced.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge