Shawn Whitfield
Virtual Cells: Predict, Explain, Discover
May 20, 2025Abstract:Drug discovery is fundamentally a process of inferring the effects of treatments on patients, and would therefore benefit immensely from computational models that can reliably simulate patient responses, enabling researchers to generate and test large numbers of therapeutic hypotheses safely and economically before initiating costly clinical trials. Even a more specific model that predicts the functional response of cells to a wide range of perturbations would be tremendously valuable for discovering safe and effective treatments that successfully translate to the clinic. Creating such virtual cells has long been a goal of the computational research community that unfortunately remains unachieved given the daunting complexity and scale of cellular biology. Nevertheless, recent advances in AI, computing power, lab automation, and high-throughput cellular profiling provide new opportunities for reaching this goal. In this perspective, we present a vision for developing and evaluating virtual cells that builds on our experience at Recursion. We argue that in order to be a useful tool to discover novel biology, virtual cells must accurately predict the functional response of a cell to perturbations and explain how the predicted response is a consequence of modifications to key biomolecular interactions. We then introduce key principles for designing therapeutically-relevant virtual cells, describe a lab-in-the-loop approach for generating novel insights with them, and advocate for biologically-grounded benchmarks to guide virtual cell development. Finally, we make the case that our approach to virtual cells provides a useful framework for building other models at higher levels of organization, including virtual patients. We hope that these directions prove useful to the research community in developing virtual models optimized for positive impact on drug discovery outcomes.
Diagnosing COVID-19 Severity from Chest X-Ray Images Using ViT and CNN Architectures
Feb 23, 2025



Abstract:The COVID-19 pandemic strained healthcare resources and prompted discussion about how machine learning can alleviate physician burdens and contribute to diagnosis. Chest x-rays (CXRs) are used for diagnosis of COVID-19, but few studies predict the severity of a patient's condition from CXRs. In this study, we produce a large COVID severity dataset by merging three sources and investigate the efficacy of transfer learning using ImageNet- and CXR-pretrained models and vision transformers (ViTs) in both severity regression and classification tasks. A pretrained DenseNet161 model performed the best on the three class severity prediction problem, reaching 80% accuracy overall and 77.3%, 83.9%, and 70% on mild, moderate and severe cases, respectively. The ViT had the best regression results, with a mean absolute error of 0.5676 compared to radiologist-predicted severity scores. The project's source code is publicly available.
Benchmarking Transcriptomics Foundation Models for Perturbation Analysis : one PCA still rules them all
Oct 17, 2024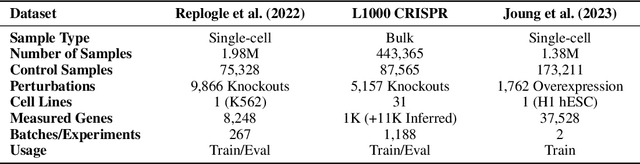
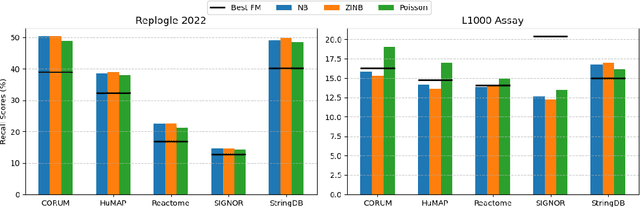
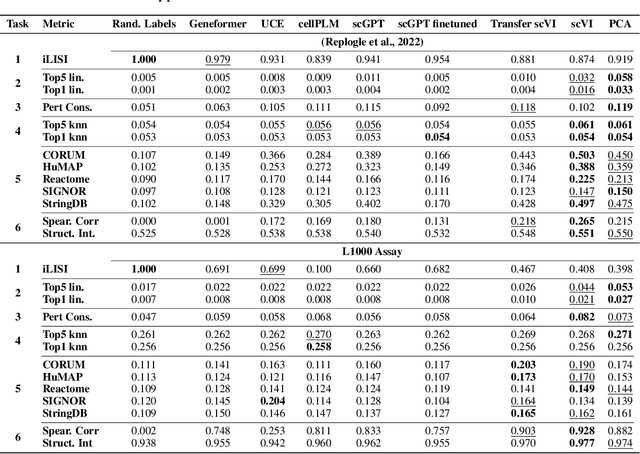
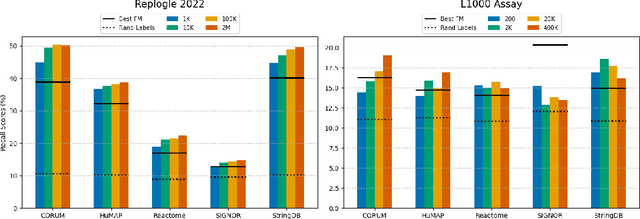
Abstract:Understanding the relationships among genes, compounds, and their interactions in living organisms remains limited due to technological constraints and the complexity of biological data. Deep learning has shown promise in exploring these relationships using various data types. However, transcriptomics, which provides detailed insights into cellular states, is still underused due to its high noise levels and limited data availability. Recent advancements in transcriptomics sequencing provide new opportunities to uncover valuable insights, especially with the rise of many new foundation models for transcriptomics, yet no benchmark has been made to robustly evaluate the effectiveness of these rising models for perturbation analysis. This article presents a novel biologically motivated evaluation framework and a hierarchy of perturbation analysis tasks for comparing the performance of pretrained foundation models to each other and to more classical techniques of learning from transcriptomics data. We compile diverse public datasets from different sequencing techniques and cell lines to assess models performance. Our approach identifies scVI and PCA to be far better suited models for understanding biological perturbations in comparison to existing foundation models, especially in their application in real-world scenarios.
Automated Discovery of Pairwise Interactions from Unstructured Data
Sep 11, 2024



Abstract:Pairwise interactions between perturbations to a system can provide evidence for the causal dependencies of the underlying underlying mechanisms of a system. When observations are low dimensional, hand crafted measurements, detecting interactions amounts to simple statistical tests, but it is not obvious how to detect interactions between perturbations affecting latent variables. We derive two interaction tests that are based on pairwise interventions, and show how these tests can be integrated into an active learning pipeline to efficiently discover pairwise interactions between perturbations. We illustrate the value of these tests in the context of biology, where pairwise perturbation experiments are frequently used to reveal interactions that are not observable from any single perturbation. Our tests can be run on unstructured data, such as the pixels in an image, which enables a more general notion of interaction than typical cell viability experiments, and can be run on cheaper experimental assays. We validate on several synthetic and real biological experiments that our tests are able to identify interacting pairs effectively. We evaluate our approach on a real biological experiment where we knocked out 50 pairs of genes and measured the effect with microscopy images. We show that we are able to recover significantly more known biological interactions than random search and standard active learning baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge