M. Rita Verdelho
The Role of Graph-based MIL and Interventional Training in the Generalization of WSI Classifiers
Jan 31, 2025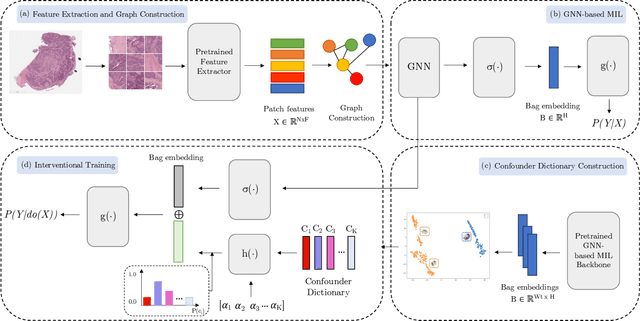



Abstract:Whole Slide Imaging (WSI), which involves high-resolution digital scans of pathology slides, has become the gold standard for cancer diagnosis, but its gigapixel resolution and the scarcity of annotated datasets present challenges for deep learning models. Multiple Instance Learning (MIL), a widely-used weakly supervised approach, bypasses the need for patch-level annotations. However, conventional MIL methods overlook the spatial relationships between patches, which are crucial for tasks such as cancer grading and diagnosis. To address this, graph-based approaches have gained prominence by incorporating spatial information through node connections. Despite their potential, both MIL and graph-based models are vulnerable to learning spurious associations, like color variations in WSIs, affecting their robustness. In this dissertation, we conduct an extensive comparison of multiple graph construction techniques, MIL models, graph-MIL approaches, and interventional training, introducing a new framework, Graph-based Multiple Instance Learning with Interventional Training (GMIL-IT), for WSI classification. We evaluate their impact on model generalization through domain shift analysis and demonstrate that graph-based models alone achieve the generalization initially anticipated from interventional training. Our code is available here: github.com/ritamartinspereira/GMIL-IT
Continual Deep Active Learning for Medical Imaging: Replay-Base Architecture for Context Adaptation
Jan 14, 2025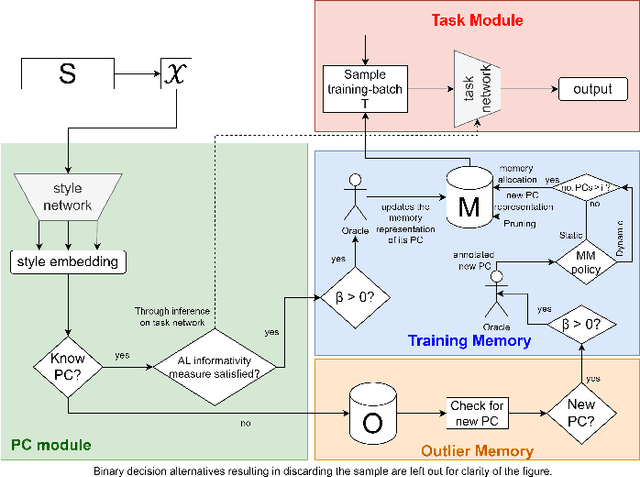
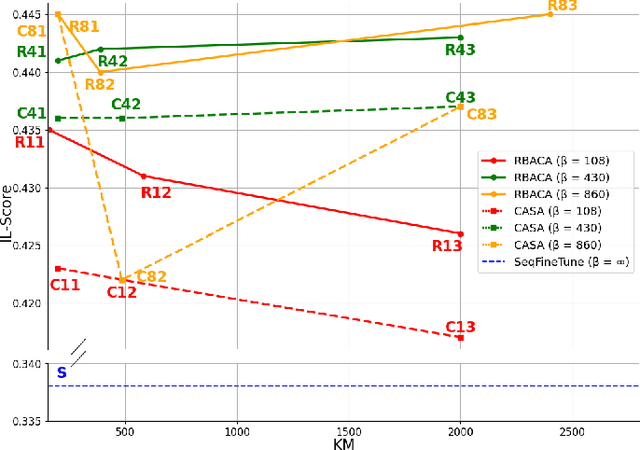
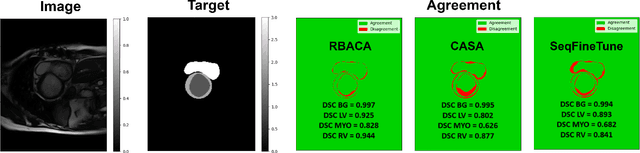
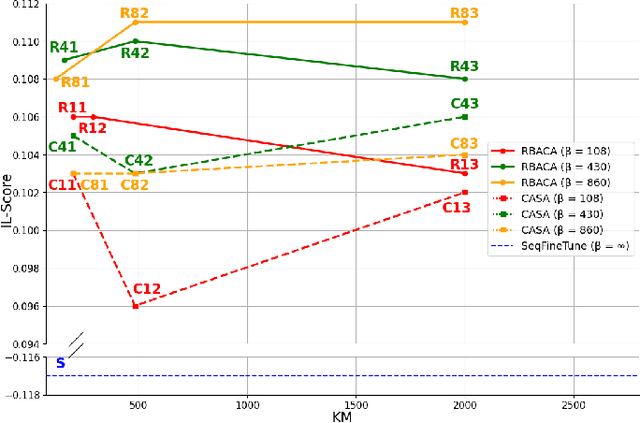
Abstract:Deep Learning for medical imaging faces challenges in adapting and generalizing to new contexts. Additionally, it often lacks sufficient labeled data for specific tasks requiring significant annotation effort. Continual Learning (CL) tackles adaptability and generalizability by enabling lifelong learning from a data stream while mitigating forgetting of previously learned knowledge. Active Learning (AL) reduces the number of required annotations for effective training. This work explores both approaches (CAL) to develop a novel framework for robust medical image analysis. Based on the automatic recognition of shifts in image characteristics, Replay-Base Architecture for Context Adaptation (RBACA) employs a CL rehearsal method to continually learn from diverse contexts, and an AL component to select the most informative instances for annotation. A novel approach to evaluate CAL methods is established using a defined metric denominated IL-Score, which allows for the simultaneous assessment of transfer learning, forgetting, and final model performance. We show that RBACA works in domain and class-incremental learning scenarios, by assessing its IL-Score on the segmentation and diagnosis of cardiac images. The results show that RBACA outperforms a baseline framework without CAL, and a state-of-the-art CAL method across various memory sizes and annotation budgets. Our code is available in https://github.com/RuiDaniel/RBACA .
MMIST-ccRCC: A Real World Medical Dataset for the Development of Multi-Modal Systems
May 02, 2024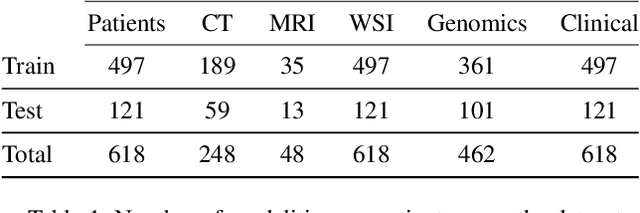
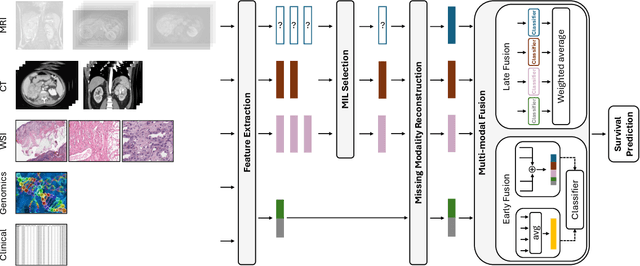
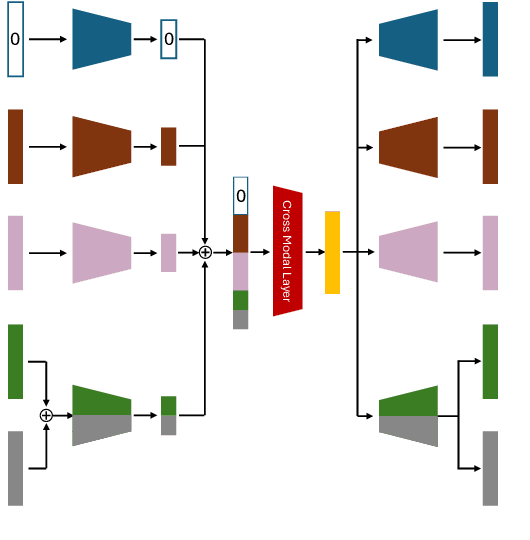
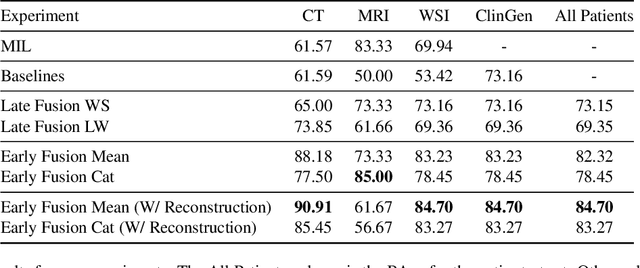
Abstract:The acquisition of different data modalities can enhance our knowledge and understanding of various diseases, paving the way for a more personalized healthcare. Thus, medicine is progressively moving towards the generation of massive amounts of multi-modal data (\emph{e.g,} molecular, radiology, and histopathology). While this may seem like an ideal environment to capitalize data-centric machine learning approaches, most methods still focus on exploring a single or a pair of modalities due to a variety of reasons: i) lack of ready to use curated datasets; ii) difficulty in identifying the best multi-modal fusion strategy; and iii) missing modalities across patients. In this paper we introduce a real world multi-modal dataset called MMIST-CCRCC that comprises 2 radiology modalities (CT and MRI), histopathology, genomics, and clinical data from 618 patients with clear cell renal cell carcinoma (ccRCC). We provide single and multi-modal (early and late fusion) benchmarks in the task of 12-month survival prediction in the challenging scenario of one or more missing modalities for each patient, with missing rates that range from 26$\%$ for genomics data to more than 90$\%$ for MRI. We show that even with such severe missing rates the fusion of modalities leads to improvements in the survival forecasting. Additionally, incorporating a strategy to generate the latent representations of the missing modalities given the available ones further improves the performance, highlighting a potential complementarity across modalities. Our dataset and code are available here: https://multi-modal-ist.github.io/datasets/ccRCC
Key Patches Are All You Need: A Multiple Instance Learning Framework For Robust Medical Diagnosis
May 02, 2024Abstract:Deep learning models have revolutionized the field of medical image analysis, due to their outstanding performances. However, they are sensitive to spurious correlations, often taking advantage of dataset bias to improve results for in-domain data, but jeopardizing their generalization capabilities. In this paper, we propose to limit the amount of information these models use to reach the final classification, by using a multiple instance learning (MIL) framework. MIL forces the model to use only a (small) subset of patches in the image, identifying discriminative regions. This mimics the clinical procedures, where medical decisions are based on localized findings. We evaluate our framework on two medical applications: skin cancer diagnosis using dermoscopy and breast cancer diagnosis using mammography. Our results show that using only a subset of the patches does not compromise diagnostic performance for in-domain data, compared to the baseline approaches. However, our approach is more robust to shifts in patient demographics, while also providing more detailed explanations about which regions contributed to the decision. Code is available at: https://github.com/diogojpa99/MedicalMultiple-Instance-Learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge