The Role of Graph-based MIL and Interventional Training in the Generalization of WSI Classifiers
Paper and Code
Jan 31, 2025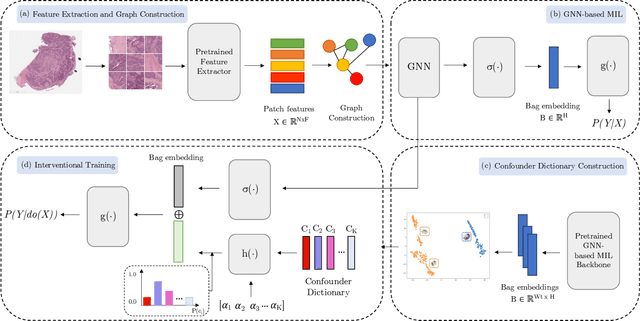



Whole Slide Imaging (WSI), which involves high-resolution digital scans of pathology slides, has become the gold standard for cancer diagnosis, but its gigapixel resolution and the scarcity of annotated datasets present challenges for deep learning models. Multiple Instance Learning (MIL), a widely-used weakly supervised approach, bypasses the need for patch-level annotations. However, conventional MIL methods overlook the spatial relationships between patches, which are crucial for tasks such as cancer grading and diagnosis. To address this, graph-based approaches have gained prominence by incorporating spatial information through node connections. Despite their potential, both MIL and graph-based models are vulnerable to learning spurious associations, like color variations in WSIs, affecting their robustness. In this dissertation, we conduct an extensive comparison of multiple graph construction techniques, MIL models, graph-MIL approaches, and interventional training, introducing a new framework, Graph-based Multiple Instance Learning with Interventional Training (GMIL-IT), for WSI classification. We evaluate their impact on model generalization through domain shift analysis and demonstrate that graph-based models alone achieve the generalization initially anticipated from interventional training. Our code is available here: github.com/ritamartinspereira/GMIL-IT
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge