Tae Hyung Kim
ZIUM: Zero-Shot Intent-Aware Adversarial Attack on Unlearned Models
Jul 29, 2025Abstract:Machine unlearning (MU) removes specific data points or concepts from deep learning models to enhance privacy and prevent sensitive content generation. Adversarial prompts can exploit unlearned models to generate content containing removed concepts, posing a significant security risk. However, existing adversarial attack methods still face challenges in generating content that aligns with an attacker's intent while incurring high computational costs to identify successful prompts. To address these challenges, we propose ZIUM, a Zero-shot Intent-aware adversarial attack on Unlearned Models, which enables the flexible customization of target attack images to reflect an attacker's intent. Additionally, ZIUM supports zero-shot adversarial attacks without requiring further optimization for previously attacked unlearned concepts. The evaluation across various MU scenarios demonstrated ZIUM's effectiveness in successfully customizing content based on user-intent prompts while achieving a superior attack success rate compared to existing methods. Moreover, its zero-shot adversarial attack significantly reduces the attack time for previously attacked unlearned concepts.
A Tutorial on MRI Reconstruction: From Modern Methods to Clinical Implications
Jul 22, 2025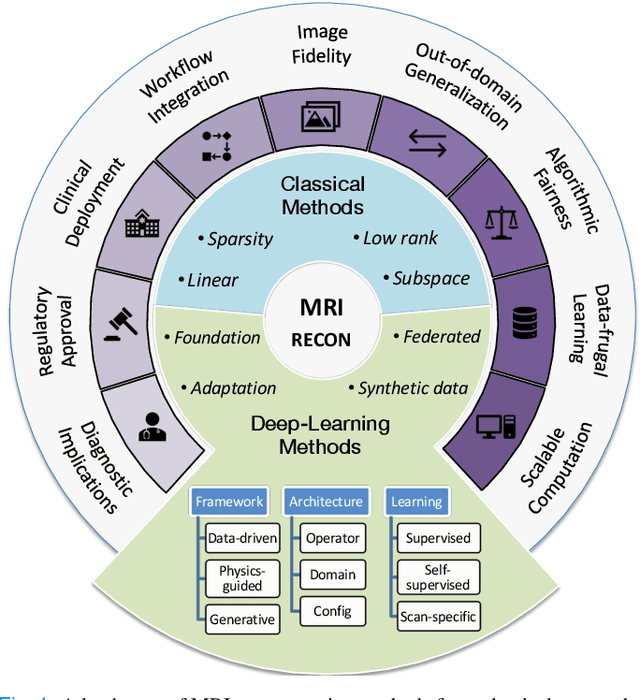
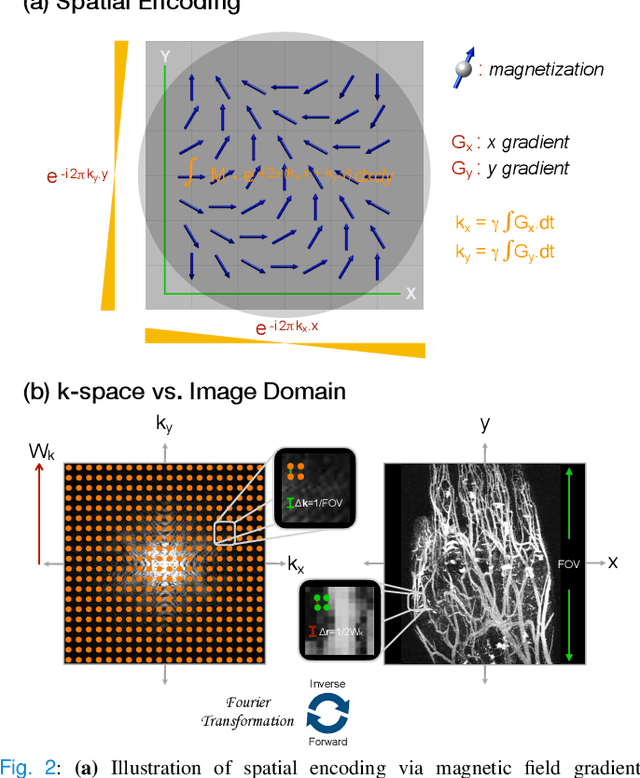
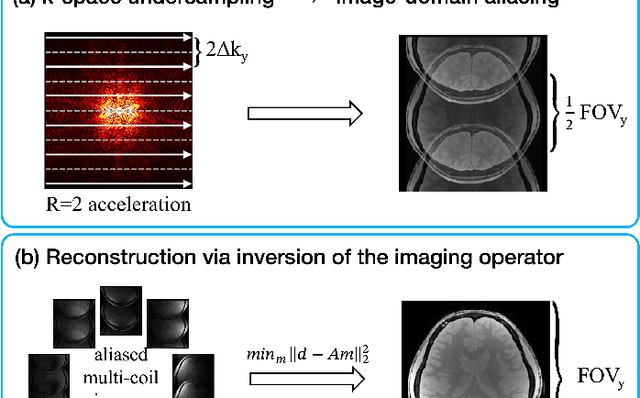
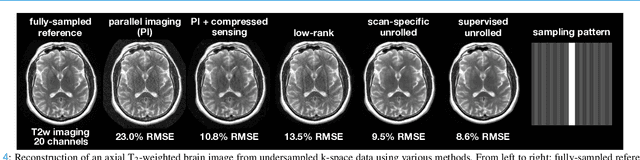
Abstract:MRI is an indispensable clinical tool, offering a rich variety of tissue contrasts to support broad diagnostic and research applications. Clinical exams routinely acquire multiple structural sequences that provide complementary information for differential diagnosis, while research protocols often incorporate advanced functional, diffusion, spectroscopic, and relaxometry sequences to capture multidimensional insights into tissue structure and composition. However, these capabilities come at the cost of prolonged scan times, which reduce patient throughput, increase susceptibility to motion artifacts, and may require trade-offs in image quality or diagnostic scope. Over the last two decades, advances in image reconstruction algorithms--alongside improvements in hardware and pulse sequence design--have made it possible to accelerate acquisitions while preserving diagnostic quality. Central to this progress is the ability to incorporate prior information to regularize the solutions to the reconstruction problem. In this tutorial, we overview the basics of MRI reconstruction and highlight state-of-the-art approaches, beginning with classical methods that rely on explicit hand-crafted priors, and then turning to deep learning methods that leverage a combination of learned and crafted priors to further push the performance envelope. We also explore the translational aspects and eventual clinical implications of these methods. We conclude by discussing future directions to address remaining challenges in MRI reconstruction. The tutorial is accompanied by a Python toolbox (https://github.com/tutorial-MRI-recon/tutorial) to demonstrate select methods discussed in the article.
Time-efficient, High Resolution 3T Whole Brain Quantitative Relaxometry using 3D-QALAS with Wave-CAIPI Readouts
Nov 08, 2022



Abstract:Purpose: Volumetric, high resolution, quantitative mapping of brain tissues relaxation properties is hindered by long acquisition times and SNR challenges. This study, for the first time, combines the time efficient wave-CAIPI readouts into the 3D-QALAS acquisition scheme, enabling full brain quantitative T1, T2 and PD maps at 1.15 isotropic voxels in only 3 minutes. Methods: Wave-CAIPI readouts were embedded in the standard 3d-QALAS encoding scheme, enabling full brain quantitative parameter maps (T1, T2 and PD) at acceleration factors of R=3x2 with minimum SNR loss due to g-factor penalties. The quantitative maps using the accelerated protocol were quantitatively compared against those obtained from conventional 3D-QALAS sequence using GRAPPA acceleration of R=2 in the ISMRM NIST phantom, and ten healthy volunteers. To show the feasibility of the proposed methods in clinical settings, the accelerated wave-CAIPI 3D-QALAS sequence was also employed in pediatric patients undergoing clinical MRI examinations. Results: When tested in both the ISMRM/NIST phantom and 7 healthy volunteers, the quantitative maps using the accelerated protocol showed excellent agreement against those obtained from conventional 3D-QALAS at R=2. Conclusion: 3D-QALAS enhanced with wave-CAIPI readouts enables time-efficient, full brain quantitative T1, T2 and PD mapping at 1.15 in 3 minutes at R=3x2 acceleration. When tested on the NIST phantom and 7 healthy volunteers, the quantitative maps obtained from the accelerated wave-CAIPI 3D-QALAS protocol showed very similar values to those obtained from the standard 3D-QALAS (R=2) protocol, alluding to the robustness and reliability of the proposed methods. This study also shows that the accelerated protocol can be effectively employed in pediatric patient populations, making high-quality high-resolution full brain quantitative imaging feasible in clinical settings.
Accurate parameter estimation using scan-specific unsupervised deep learning for relaxometry and MR fingerprinting
Dec 13, 2021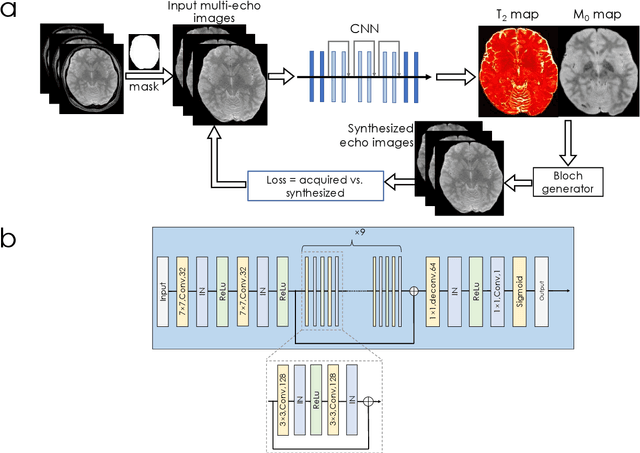
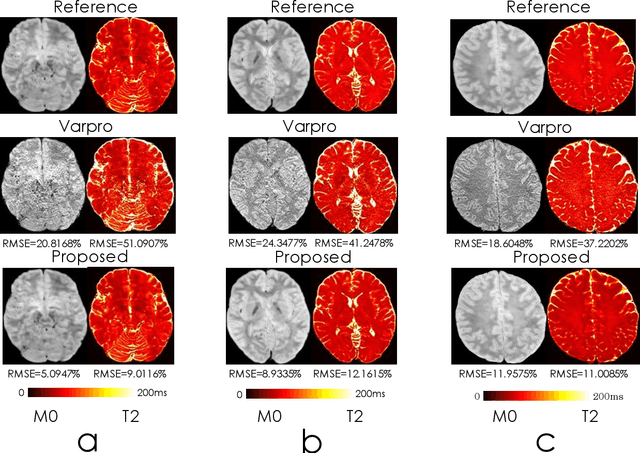
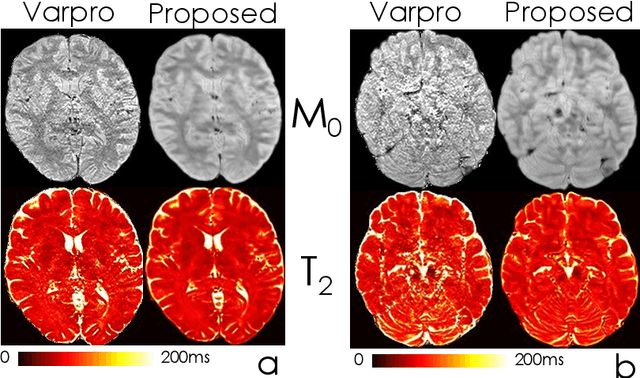
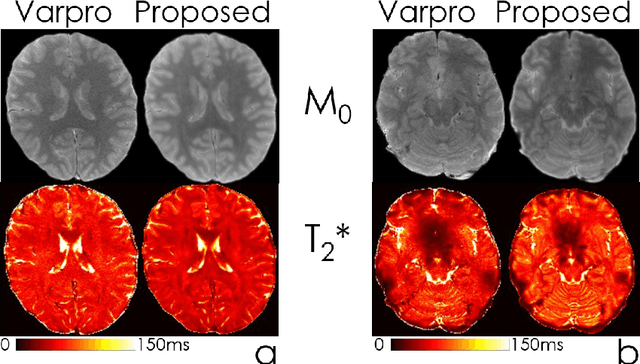
Abstract:We propose an unsupervised convolutional neural network (CNN) for relaxation parameter estimation. This network incorporates signal relaxation and Bloch simulations while taking advantage of residual learning and spatial relations across neighboring voxels. Quantification accuracy and robustness to noise is shown to be significantly improved compared to standard parameter estimation methods in numerical simulations and in vivo data for multi-echo T2 and T2* mapping. The combination of the proposed network with subspace modeling and MR fingerprinting (MRF) from highly undersampled data permits high quality T1 and T2 mapping.
LGSVL Simulator: A High Fidelity Simulator for Autonomous Driving
May 11, 2020
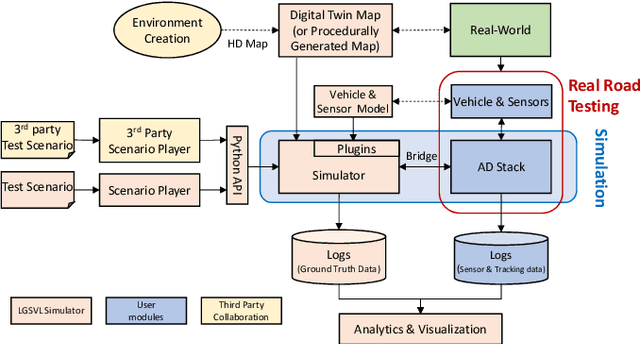
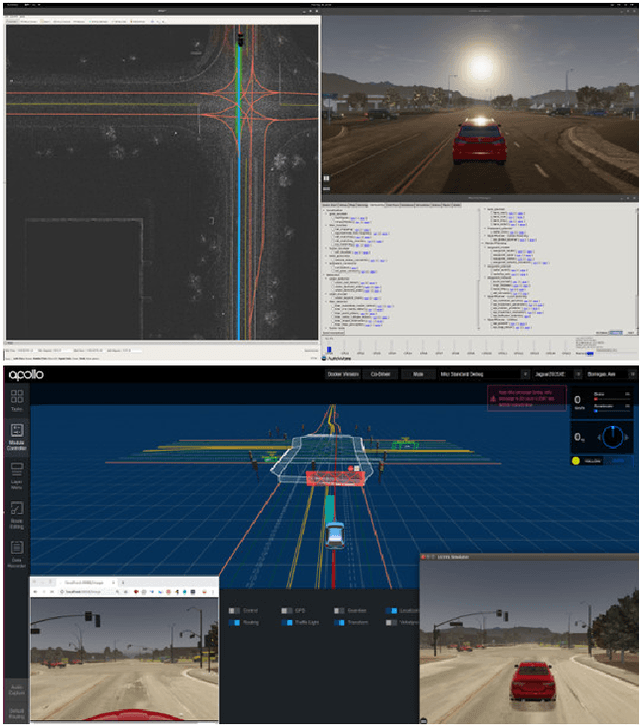
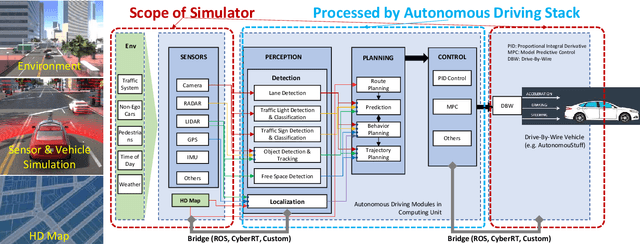
Abstract:Testing autonomous driving algorithms on real autonomous vehicles is extremely costly and many researchers and developers in the field cannot afford a real car and the corresponding sensors. Although several free and open-source autonomous driving stacks, such as Autoware and Apollo are available, choices of open-source simulators to use with them are limited. In this paper, we introduce the LGSVL Simulator which is a high fidelity simulator for autonomous driving. The simulator engine provides end-to-end, full-stack simulation which is ready to be hooked up to Autoware and Apollo. In addition, simulator tools are provided with the core simulation engine which allow users to easily customize sensors, create new types of controllable objects, replace some modules in the core simulator, and create digital twins of particular environments.
LORAKI: Autocalibrated Recurrent Neural Networks for Autoregressive MRI Reconstruction in k-Space
Apr 24, 2019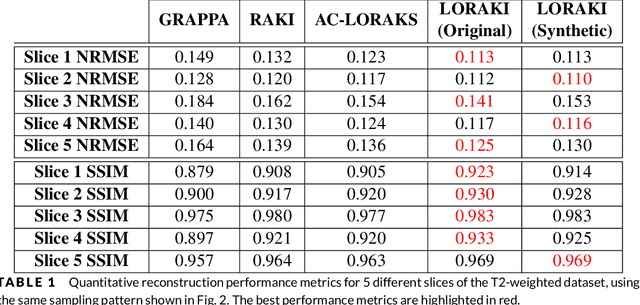


Abstract:We propose and evaluate a new MRI reconstruction method named LORAKI that trains an autocalibrated scan-specific recurrent neural network (RNN) to recover missing k-space data. Methods like GRAPPA, SPIRiT, and AC-LORAKS assume that k-space data has shift-invariant autoregressive structure, and that the scan-specific autoregression relationships needed to recover missing samples can be learned from fully-sampled autocalibration (ACS) data. Recently, the structure of the linear GRAPPA method has been translated into a nonlinear deep learning method named RAKI. RAKI uses ACS data to train an artificial neural network to interpolate missing k-space samples, and often outperforms GRAPPA. In this work, we apply a similar principle to translate the linear AC-LORAKS method (simultaneously incorporating support, phase, and parallel imaging constraints) into a nonlinear deep learning method named LORAKI. Since AC-LORAKS is iterative and convolutional, LORAKI takes the form of a convolutional RNN. This new architecture admits a wide range of sampling patterns, and even calibrationless patterns are possible if synthetic ACS data is generated. The performance of LORAKI was evaluated with retrospectively undersampled brain datasets, with comparisons against other related reconstruction methods. Results suggest that LORAKI can provide improved reconstruction compared to other scan-specific autocalibrated reconstruction methods like GRAPPA, RAKI, and AC-LORAKS. LORAKI offers a new deep-learning approach to MRI reconstruction based on RNNs in k-space, and enables improved image quality and enhanced sampling flexibility.
Navigator-free EPI Ghost Correction with Structured Low-Rank Matrix Models: New Theory and Methods
Mar 06, 2018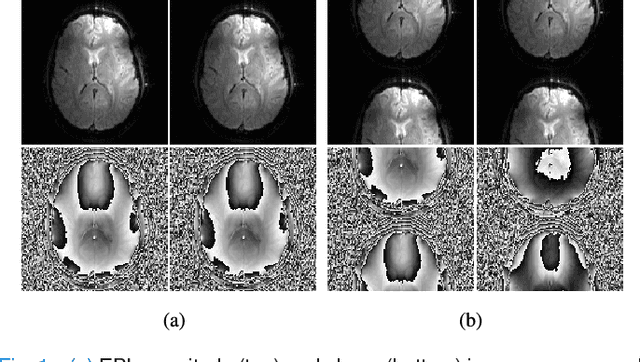


Abstract:Structured low-rank matrix models have previously been introduced to enable calibrationless MR image reconstruction from sub-Nyquist data, and such ideas have recently been extended to enable navigator-free echo-planar imaging (EPI) ghost correction. This paper presents novel theoretical analysis which shows that, because of uniform subsampling, the structured low-rank matrix optimization problems for EPI data will always have either undesirable or non-unique solutions in the absence of additional constraints. This theory leads us to recommend and investigate problem formulations for navigator-free EPI that incorporate side information from either image-domain or k-space domain parallel imaging methods. The importance of using nonconvex low-rank matrix regularization is also identified. We demonstrate using phantom and \emph{in vivo} data that the proposed methods are able to eliminate ghost artifacts for several navigator-free EPI acquisition schemes, obtaining better performance in comparison to state-of-the-art methods across a range of different scenarios. Results are shown for both single-channel acquisition and highly accelerated multi-channel acquisition.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge