Olivio Donati
Constrained Optimization for Training Deep Neural Networks Under Class Imbalance
Feb 21, 2021
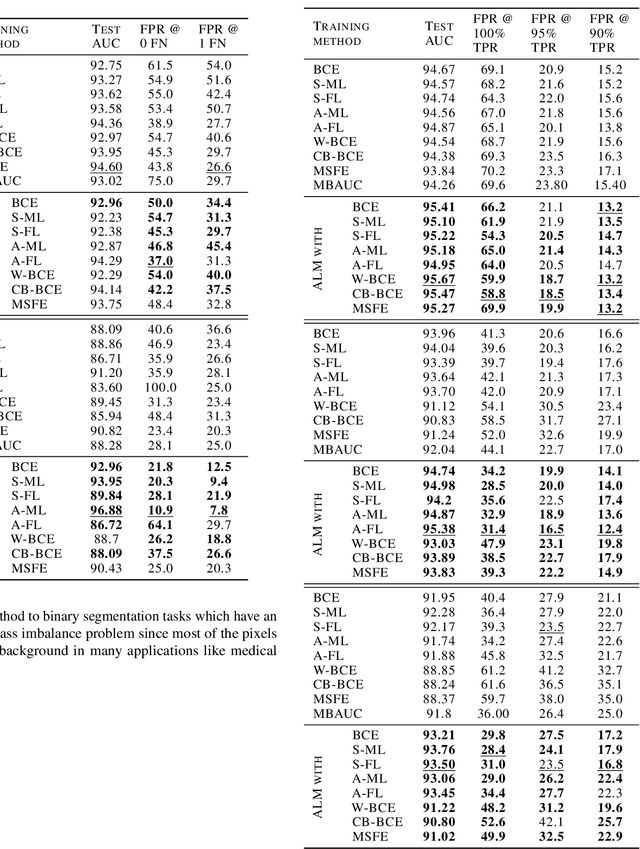

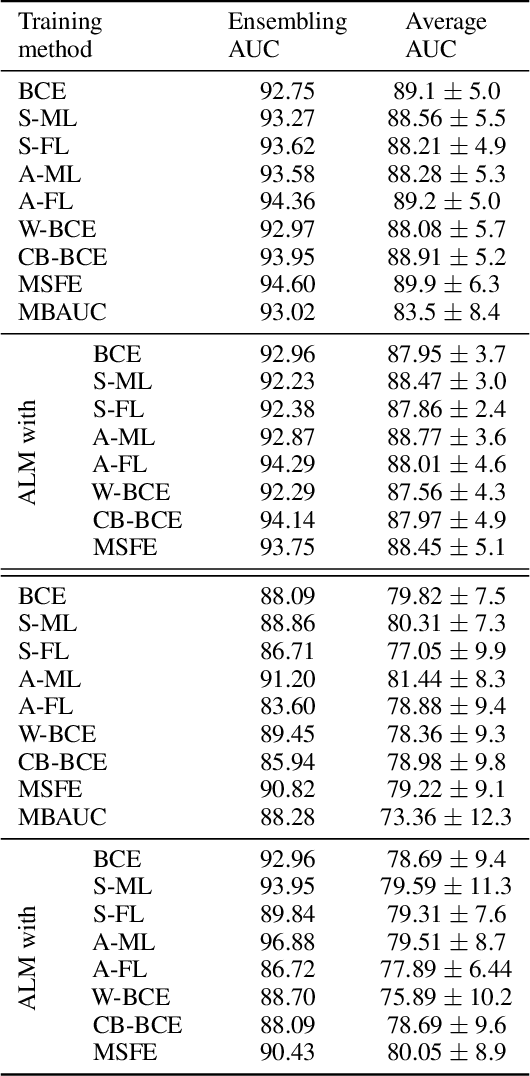
Abstract:Deep neural networks (DNNs) are notorious for making more mistakes for the classes that have substantially fewer samples than the others during training. Such class imbalance is ubiquitous in clinical applications and very crucial to handle because the classes with fewer samples most often correspond to critical cases (e.g., cancer) where misclassifications can have severe consequences. Not to miss such cases, binary classifiers need to be operated at high True Positive Rates (TPR) by setting a higher threshold but this comes at the cost of very high False Positive Rates (FPR) for problems with class imbalance. Existing methods for learning under class imbalance most often do not take this into account. We argue that prediction accuracy should be improved by emphasizing reducing FPRs at high TPRs for problems where misclassification of the positive samples are associated with higher cost. To this end, we pose the training of a DNN for binary classification as a constrained optimization problem and introduce a novel constraint that can be used with existing loss functions to enforce maximal area under the ROC curve (AUC). We solve the resulting constrained optimization problem using an Augmented Lagrangian method (ALM), where the constraint emphasizes reduction of FPR at high TPR. We present experimental results for image-based classification applications using the CIFAR10 and an in-house medical imaging dataset. Our results demonstrate that the proposed method almost always improves the loss functions it is used with by attaining lower FPR at high TPR and higher or equal AUC.
Semi-supervised Task-driven Data Augmentation for Medical Image Segmentation
Jul 09, 2020



Abstract:Supervised learning-based segmentation methods typically require a large number of annotated training data to generalize well at test time. In medical applications, curating such datasets is not a favourable option because acquiring a large number of annotated samples from experts is time-consuming and expensive. Consequently, numerous methods have been proposed in the literature for learning with limited annotated examples. Unfortunately, the proposed approaches in the literature have not yet yielded significant gains over random data augmentation for image segmentation, where random augmentations themselves do not yield high accuracy. In this work, we propose a novel task-driven data augmentation method for learning with limited labeled data where the synthetic data generator, is optimized for the segmentation task. The generator of the proposed method models intensity and shape variations using two sets of transformations, as additive intensity transformations and deformation fields. Both transformations are optimized using labeled as well as unlabeled examples in a semi-supervised framework. Our experiments on three medical datasets, namely cardic, prostate and pancreas, show that the proposed approach significantly outperforms standard augmentation and semi-supervised approaches for image segmentation in the limited annotation setting. The code is made publicly available at https://github.com/krishnabits001/task$\_$driven$\_$data$\_$augmentation.
PHiSeg: Capturing Uncertainty in Medical Image Segmentation
Jun 07, 2019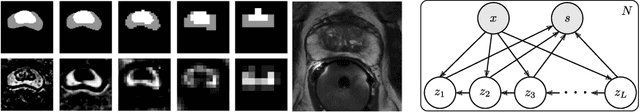

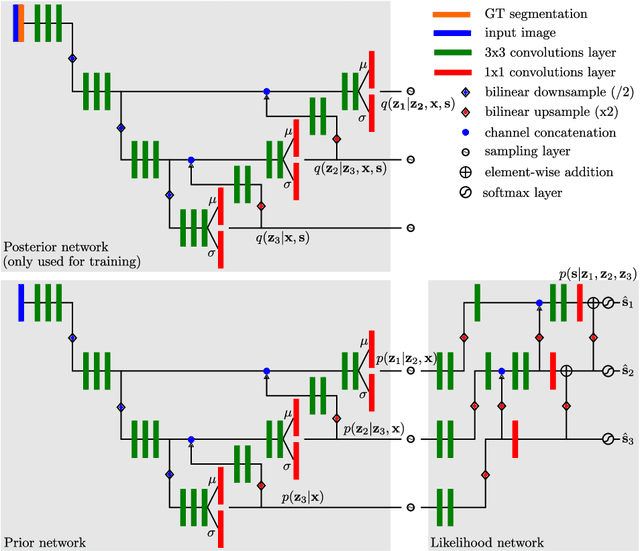
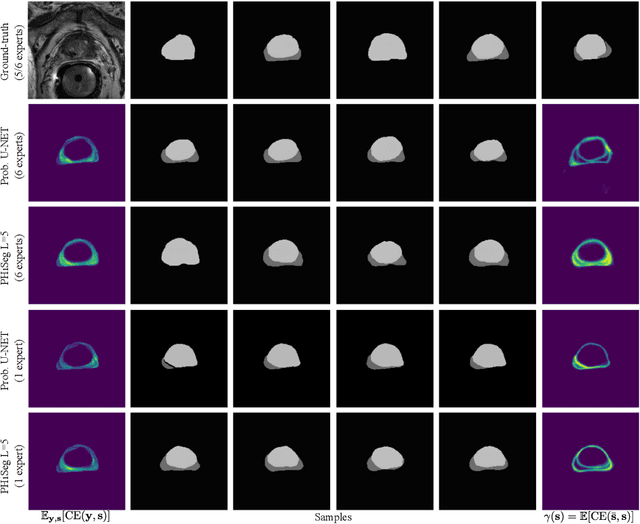
Abstract:Segmentation of anatomical structures and pathologies is inherently ambiguous. For instance, structure borders may not be clearly visible or different experts may have different styles of annotating. The majority of current state-of-the-art methods do not account for such ambiguities but rather learn a single mapping from image to segmentation. In this work, we propose a novel method to model the conditional probability distribution of the segmentations given an input image. We derive a hierarchical probabilistic model, in which separate latent spaces are responsible for modelling the segmentation at different resolutions. Inference in this model can be efficiently performed using the variational autoencoder framework. We show that our proposed method can be used to generate significantly more realistic and diverse segmentation samples compared to recent related work, both, when trained with annotations from a single or multiple annotators.
Semi-Supervised and Task-Driven Data Augmentation
Feb 28, 2019



Abstract:Supervised deep learning methods for segmentation require large amounts of labelled training data, without which they are prone to overfitting, not generalizing well to unseen images. In practice, obtaining a large number of annotations from clinical experts is expensive and time-consuming. One way to address scarcity of annotated examples is data augmentation using random spatial and intensity transformations. Recently, it has been proposed to use generative models to synthesize realistic training examples, complementing the random augmentation. So far, these methods have yielded limited gains over the random augmentation. However, there is potential to improve the approach by (i) explicitly modeling deformation fields (non-affine spatial transformation) and intensity transformations and (ii) leveraging unlabelled data during the generative process. With this motivation, we propose a novel task-driven data augmentation method where to synthesize new training examples, a generative network explicitly models and applies deformation fields and additive intensity masks on existing labelled data, modeling shape and intensity variations, respectively. Crucially, the generative model is optimized to be conducive to the task, in this case segmentation, and constrained to match the distribution of images observed from labelled and unlabelled samples. Furthermore, explicit modeling of deformation fields allow synthesizing segmentation masks and images in exact correspondence by simply applying the generated transformation to an input image and the corresponding annotation. Our experiments on cardiac magnetic resonance images (MRI) showed that, for the task of segmentation in small training data scenarios, the proposed method substantially outperforms conventional augmentation techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge