Anton S. Becker
PHiSeg: Capturing Uncertainty in Medical Image Segmentation
Jun 07, 2019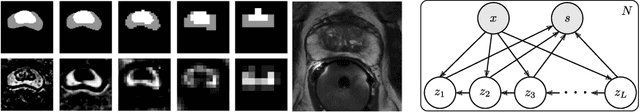

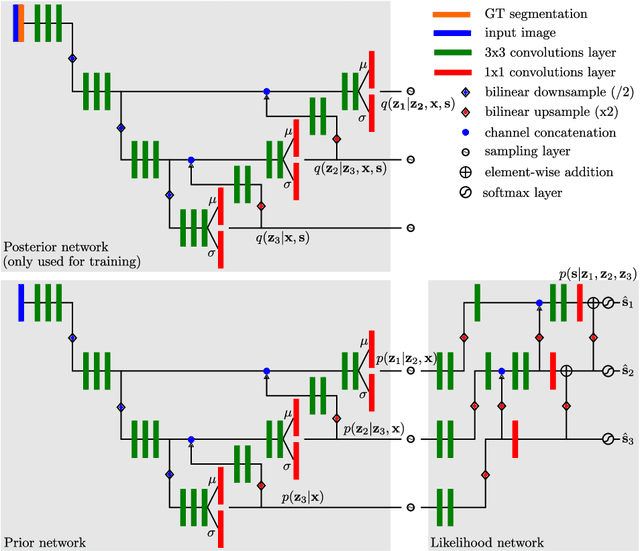
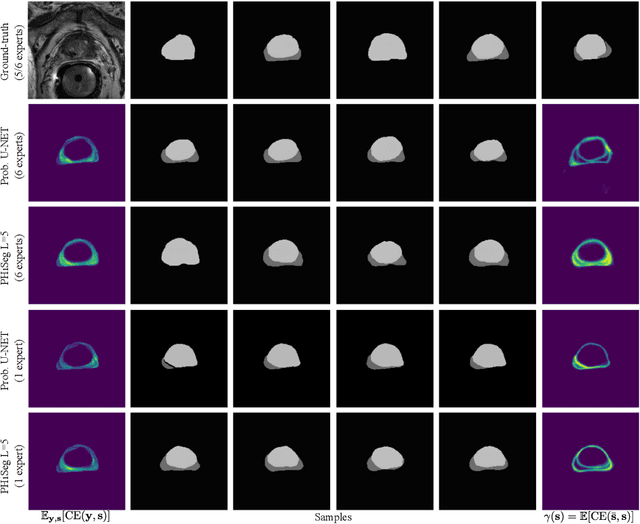
Abstract:Segmentation of anatomical structures and pathologies is inherently ambiguous. For instance, structure borders may not be clearly visible or different experts may have different styles of annotating. The majority of current state-of-the-art methods do not account for such ambiguities but rather learn a single mapping from image to segmentation. In this work, we propose a novel method to model the conditional probability distribution of the segmentations given an input image. We derive a hierarchical probabilistic model, in which separate latent spaces are responsible for modelling the segmentation at different resolutions. Inference in this model can be efficiently performed using the variational autoencoder framework. We show that our proposed method can be used to generate significantly more realistic and diverse segmentation samples compared to recent related work, both, when trained with annotations from a single or multiple annotators.
Adversarial Augmentation for Enhancing Classification of Mammography Images
Feb 20, 2019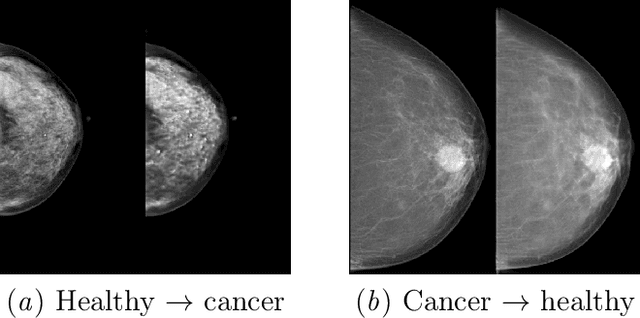
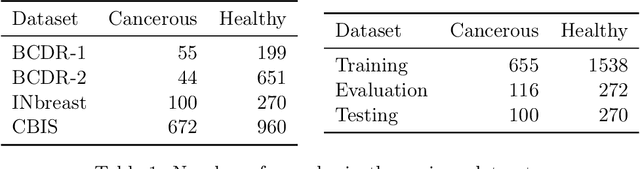
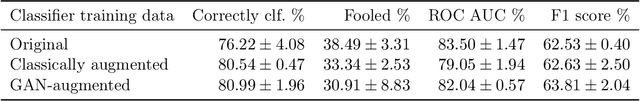
Abstract:Supervised deep learning relies on the assumption that enough training data is available, which presents a problem for its application to several fields, like medical imaging. On the example of a binary image classification task (breast cancer recognition), we show that pretraining a generative model for meaningful image augmentation helps enhance the performance of the resulting classifier. By augmenting the data, performance on downstream classification tasks could be improved even with a relatively small training set. We show that this "adversarial augmentation" yields promising results compared to classical image augmentation on the example of breast cancer classification.
Injecting and removing malignant features in mammography with CycleGAN: Investigation of an automated adversarial attack using neural networks
Nov 19, 2018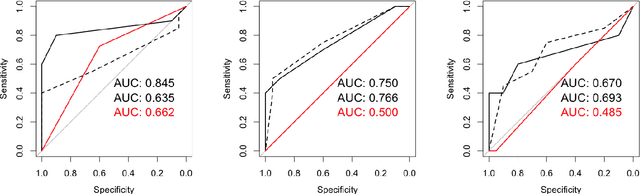
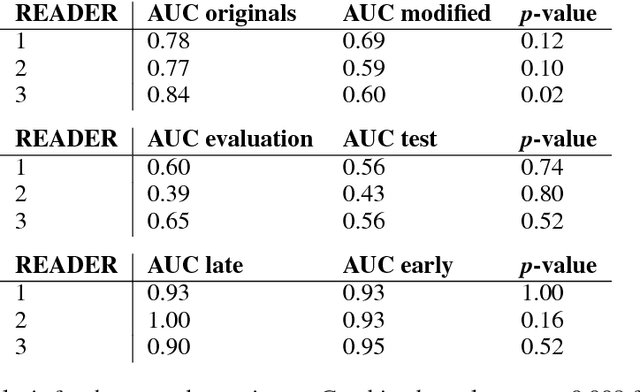

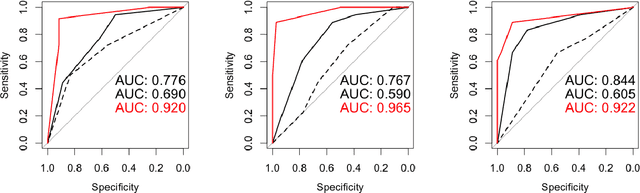
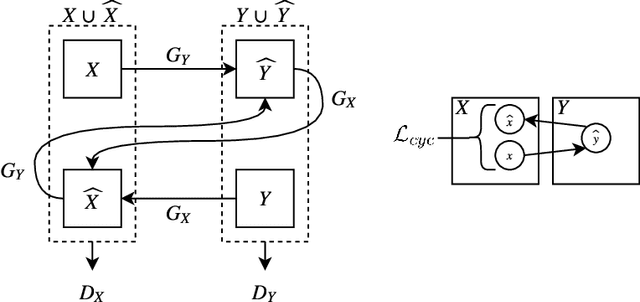
Abstract:$\textbf{Purpose}$ To train a cycle-consistent generative adversarial network (CycleGAN) on mammographic data to inject or remove features of malignancy, and to determine whether these AI-mediated attacks can be detected by radiologists. $\textbf{Material and Methods}$ From the two publicly available datasets, BCDR and INbreast, we selected images from cancer patients and healthy controls. An internal dataset served as test data, withheld during training. We ran two experiments training CycleGAN on low and higher resolution images ($256 \times 256$ px and $512 \times 408$ px). Three radiologists read the images and rated the likelihood of malignancy on a scale from 1-5 and the likelihood of the image being manipulated. The readout was evaluated by ROC analysis (Area under the ROC curve = AUC). $\textbf{Results}$ At the lower resolution, only one radiologist exhibited markedly lower detection of cancer (AUC=0.85 vs 0.63, p=0.06), while the other two were unaffected (0.67 vs. 0.69 and 0.75 vs. 0.77, p=0.55). Only one radiologist could discriminate between original and modified images slightly better than guessing/chance (0.66, p=0.008). At the higher resolution, all radiologists showed significantly lower detection rate of cancer in the modified images (0.77-0.84 vs. 0.59-0.69, p=0.008), however, they were now able to reliably detect modified images due to better visibility of artifacts (0.92, 0.92 and 0.97). $\textbf{Conclusion}$ A CycleGAN can implicitly learn malignant features and inject or remove them so that a substantial proportion of small mammographic images would consequently be misdiagnosed. At higher resolutions, however, the method is currently limited and has a clear trade-off between manipulation of images and introduction of artifacts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge