Ertunc Erdil
Conformal forecasting for surgical instrument trajectory
Mar 06, 2025


Abstract:Forecasting surgical instrument trajectories and predicting the next surgical action recently started to attract attention from the research community. Both these tasks are crucial for automation and assistance in endoscopy surgery. Given the safety-critical nature of these tasks, reliable uncertainty quantification is essential. Conformal prediction is a fast-growing and widely recognized framework for uncertainty estimation in machine learning and computer vision, offering distribution-free, theoretically valid prediction intervals. In this work, we explore the application of standard conformal prediction and conformalized quantile regression to estimate uncertainty in forecasting surgical instrument motion, i.e., predicting direction and magnitude of surgical instruments' future motion. We analyze and compare their coverage and interval sizes, assessing the impact of multiple hypothesis testing and correction methods. Additionally, we show how these techniques can be employed to produce useful uncertainty heatmaps. To the best of our knowledge, this is the first study applying conformal prediction to surgical guidance, marking an initial step toward constructing principled prediction intervals with formal coverage guarantees in this domain.
Do Vision Foundation Models Enhance Domain Generalization in Medical Image Segmentation?
Sep 12, 2024Abstract:Neural networks achieve state-of-the-art performance in many supervised learning tasks when the training data distribution matches the test data distribution. However, their performance drops significantly under domain (covariate) shift, a prevalent issue in medical image segmentation due to varying acquisition settings across different scanner models and protocols. Recently, foundational models (FMs) trained on large datasets have gained attention for their ability to be adapted for downstream tasks and achieve state-of-the-art performance with excellent generalization capabilities on natural images. However, their effectiveness in medical image segmentation remains underexplored. In this paper, we investigate the domain generalization performance of various FMs, including DinoV2, SAM, MedSAM, and MAE, when fine-tuned using various parameter-efficient fine-tuning (PEFT) techniques such as Ladder and Rein (+LoRA) and decoder heads. We introduce a novel decode head architecture, HQHSAM, which simply integrates elements from two state-of-the-art decoder heads, HSAM and HQSAM, to enhance segmentation performance. Our extensive experiments on multiple datasets, encompassing various anatomies and modalities, reveal that FMs, particularly with the HQHSAM decode head, improve domain generalization for medical image segmentation. Moreover, we found that the effectiveness of PEFT techniques varies across different FMs. These findings underscore the potential of FMs to enhance the domain generalization performance of neural networks in medical image segmentation across diverse clinical settings, providing a solid foundation for future research. Code and models are available for research purposes at \url{https://github.com/kerem-cekmeceli/Foundation-Models-for-Medical-Imagery}.
Expert load matters: operating networks at high accuracy and low manual effort
Aug 09, 2023Abstract:In human-AI collaboration systems for critical applications, in order to ensure minimal error, users should set an operating point based on model confidence to determine when the decision should be delegated to human experts. Samples for which model confidence is lower than the operating point would be manually analysed by experts to avoid mistakes. Such systems can become truly useful only if they consider two aspects: models should be confident only for samples for which they are accurate, and the number of samples delegated to experts should be minimized. The latter aspect is especially crucial for applications where available expert time is limited and expensive, such as healthcare. The trade-off between the model accuracy and the number of samples delegated to experts can be represented by a curve that is similar to an ROC curve, which we refer to as confidence operating characteristic (COC) curve. In this paper, we argue that deep neural networks should be trained by taking into account both accuracy and expert load and, to that end, propose a new complementary loss function for classification that maximizes the area under this COC curve. This promotes simultaneously the increase in network accuracy and the reduction in number of samples delegated to humans. We perform experiments on multiple computer vision and medical image datasets for classification. Our results demonstrate that the proposed loss improves classification accuracy and delegates less number of decisions to experts, achieves better out-of-distribution samples detection and on par calibration performance compared to existing loss functions.
Explicitly Minimizing the Blur Error of Variational Autoencoders
Apr 12, 2023



Abstract:Variational autoencoders (VAEs) are powerful generative modelling methods, however they suffer from blurry generated samples and reconstructions compared to the images they have been trained on. Significant research effort has been spent to increase the generative capabilities by creating more flexible models but often flexibility comes at the cost of higher complexity and computational cost. Several works have focused on altering the reconstruction term of the evidence lower bound (ELBO), however, often at the expense of losing the mathematical link to maximizing the likelihood of the samples under the modeled distribution. Here we propose a new formulation of the reconstruction term for the VAE that specifically penalizes the generation of blurry images while at the same time still maximizing the ELBO under the modeled distribution. We show the potential of the proposed loss on three different data sets, where it outperforms several recently proposed reconstruction losses for VAEs.
A Field of Experts Prior for Adapting Neural Networks at Test Time
Feb 10, 2022



Abstract:Performance of convolutional neural networks (CNNs) in image analysis tasks is often marred in the presence of acquisition-related distribution shifts between training and test images. Recently, it has been proposed to tackle this problem by fine-tuning trained CNNs for each test image. Such test-time-adaptation (TTA) is a promising and practical strategy for improving robustness to distribution shifts as it requires neither data sharing between institutions nor annotating additional data. Previous TTA methods use a helper model to increase similarity between outputs and/or features extracted from a test image with those of the training images. Such helpers, which are typically modeled using CNNs, can be task-specific and themselves vulnerable to distribution shifts in their inputs. To overcome these problems, we propose to carry out TTA by matching the feature distributions of test and training images, as modelled by a field-of-experts (FoE) prior. FoEs model complicated probability distributions as products of many simpler expert distributions. We use 1D marginal distributions of a trained task CNN's features as experts in the FoE model. Further, we compute principal components of patches of the task CNN's features, and consider the distributions of PCA loadings as additional experts. We validate the method on 5 MRI segmentation tasks (healthy tissues in 4 anatomical regions and lesions in 1 one anatomy), using data from 17 clinics, and on a MRI registration task, using data from 3 clinics. We find that the proposed FoE-based TTA is generically applicable in multiple tasks, and outperforms all previous TTA methods for lesion segmentation. For healthy tissue segmentation, the proposed method outperforms other task-agnostic methods, but a previous TTA method which is specifically designed for segmentation performs the best for most of the tested datasets. Our code is publicly available.
Wiener Guided DIP for Unsupervised Blind Image Deconvolution
Dec 19, 2021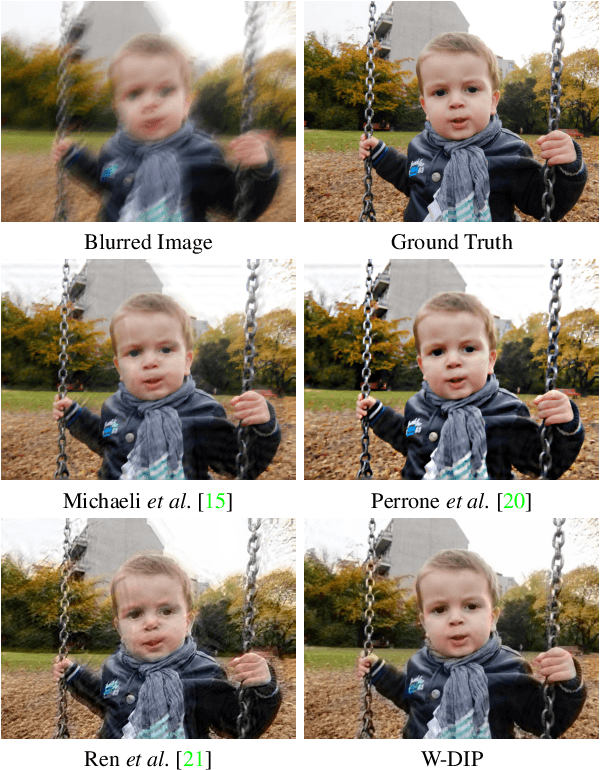
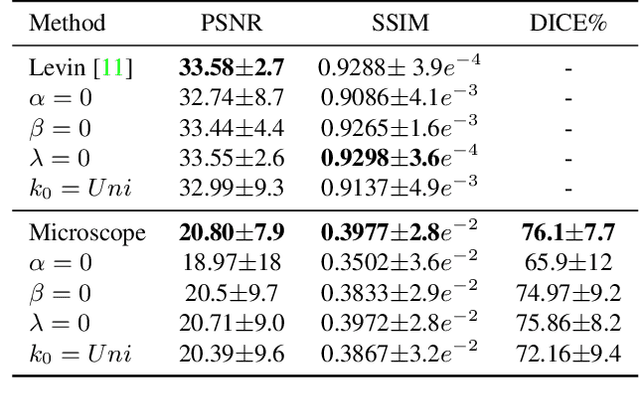
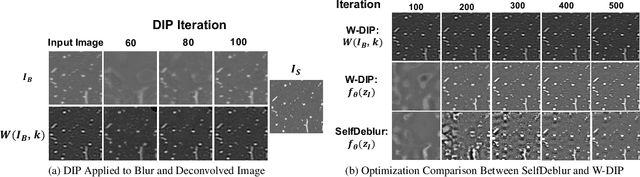
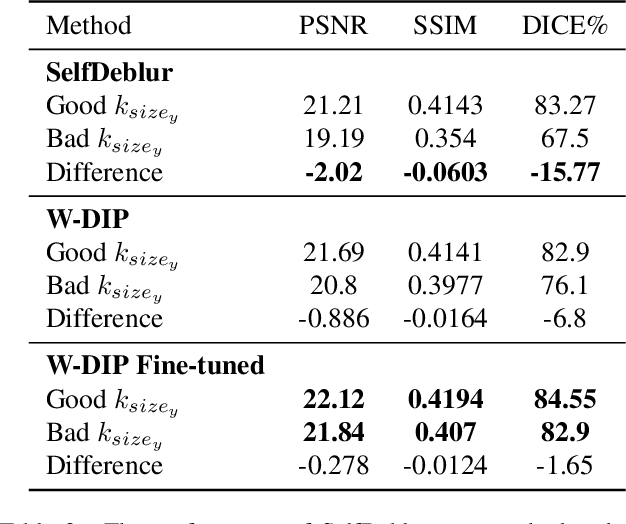
Abstract:Blind deconvolution is an ill-posed problem arising in various fields ranging from microscopy to astronomy. The ill-posed nature of the problem requires adequate priors to arrive to a desirable solution. Recently, it has been shown that deep learning architectures can serve as an image generation prior during unsupervised blind deconvolution optimization, however often exhibiting a performance fluctuation even on a single image. We propose to use Wiener-deconvolution to guide the image generator during optimization by providing it a sharpened version of the blurry image using an auxiliary kernel estimate starting from a Gaussian. We observe that the high-frequency artifacts of deconvolution are reproduced with a delay compared to low-frequency features. In addition, the image generator reproduces low-frequency features of the deconvolved image faster than that of a blurry image. We embed the computational process in a constrained optimization framework and show that the proposed method yields higher stability and performance across multiple datasets. In addition, we provide the code.
Local contrastive loss with pseudo-label based self-training for semi-supervised medical image segmentation
Dec 17, 2021



Abstract:Supervised deep learning-based methods yield accurate results for medical image segmentation. However, they require large labeled datasets for this, and obtaining them is a laborious task that requires clinical expertise. Semi/self-supervised learning-based approaches address this limitation by exploiting unlabeled data along with limited annotated data. Recent self-supervised learning methods use contrastive loss to learn good global level representations from unlabeled images and achieve high performance in classification tasks on popular natural image datasets like ImageNet. In pixel-level prediction tasks such as segmentation, it is crucial to also learn good local level representations along with global representations to achieve better accuracy. However, the impact of the existing local contrastive loss-based methods remains limited for learning good local representations because similar and dissimilar local regions are defined based on random augmentations and spatial proximity; not based on the semantic label of local regions due to lack of large-scale expert annotations in the semi/self-supervised setting. In this paper, we propose a local contrastive loss to learn good pixel level features useful for segmentation by exploiting semantic label information obtained from pseudo-labels of unlabeled images alongside limited annotated images. In particular, we define the proposed loss to encourage similar representations for the pixels that have the same pseudo-label/ label while being dissimilar to the representation of pixels with different pseudo-label/label in the dataset. We perform pseudo-label based self-training and train the network by jointly optimizing the proposed contrastive loss on both labeled and unlabeled sets and segmentation loss on only the limited labeled set. We evaluated on three public cardiac and prostate datasets, and obtain high segmentation performance.
Constrained Optimization for Training Deep Neural Networks Under Class Imbalance
Feb 21, 2021
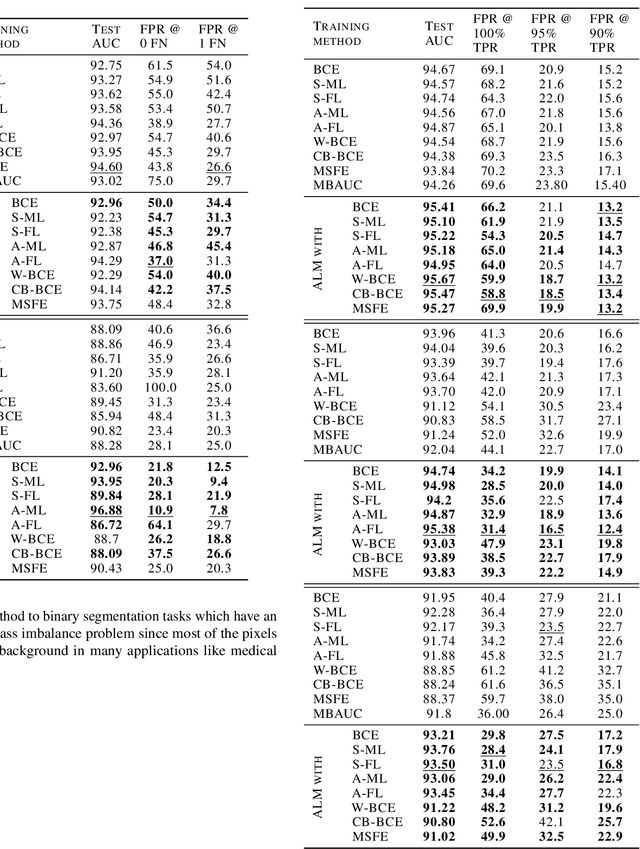

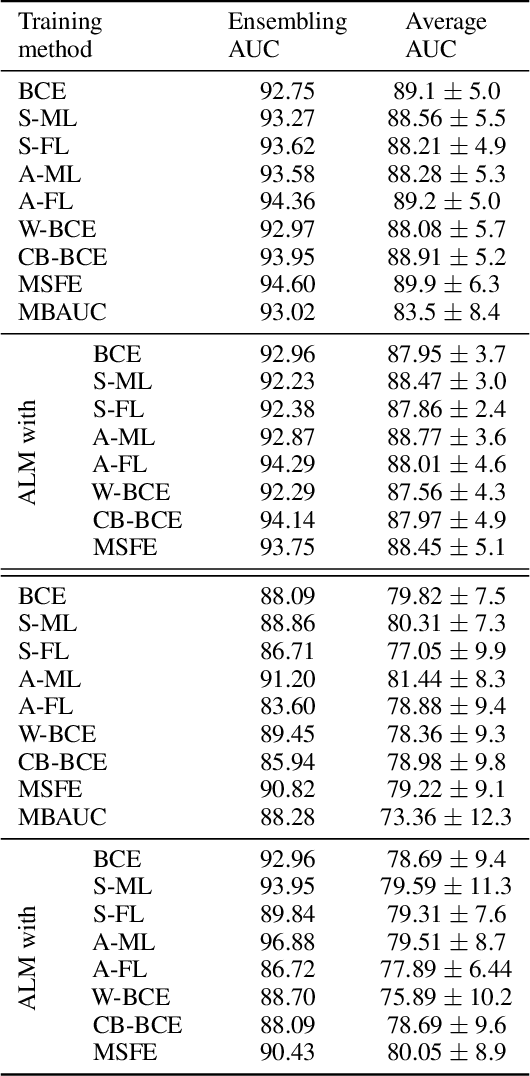
Abstract:Deep neural networks (DNNs) are notorious for making more mistakes for the classes that have substantially fewer samples than the others during training. Such class imbalance is ubiquitous in clinical applications and very crucial to handle because the classes with fewer samples most often correspond to critical cases (e.g., cancer) where misclassifications can have severe consequences. Not to miss such cases, binary classifiers need to be operated at high True Positive Rates (TPR) by setting a higher threshold but this comes at the cost of very high False Positive Rates (FPR) for problems with class imbalance. Existing methods for learning under class imbalance most often do not take this into account. We argue that prediction accuracy should be improved by emphasizing reducing FPRs at high TPRs for problems where misclassification of the positive samples are associated with higher cost. To this end, we pose the training of a DNN for binary classification as a constrained optimization problem and introduce a novel constraint that can be used with existing loss functions to enforce maximal area under the ROC curve (AUC). We solve the resulting constrained optimization problem using an Augmented Lagrangian method (ALM), where the constraint emphasizes reduction of FPR at high TPR. We present experimental results for image-based classification applications using the CIFAR10 and an in-house medical imaging dataset. Our results demonstrate that the proposed method almost always improves the loss functions it is used with by attaining lower FPR at high TPR and higher or equal AUC.
RevPHiSeg: A Memory-Efficient Neural Network for Uncertainty Quantification in Medical Image Segmentation
Aug 18, 2020



Abstract:Quantifying segmentation uncertainty has become an important issue in medical image analysis due to the inherent ambiguity of anatomical structures and its pathologies. Recently, neural network-based uncertainty quantification methods have been successfully applied to various problems. One of the main limitations of the existing techniques is the high memory requirement during training; which limits their application to processing smaller field-of-views (FOVs) and/or using shallower architectures. In this paper, we investigate the effect of using reversible blocks for building memory-efficient neural network architectures for quantification of segmentation uncertainty. The reversible architecture achieves memory saving by exactly computing the activations from the outputs of the subsequent layers during backpropagation instead of storing the activations for each layer. We incorporate the reversible blocks into a recently proposed architecture called PHiSeg that is developed for uncertainty quantification in medical image segmentation. The reversible architecture, RevPHiSeg, allows training neural networks for quantifying segmentation uncertainty on GPUs with limited memory and processing larger FOVs. We perform experiments on the LIDC-IDRI dataset and an in-house prostate dataset, and present comparisons with PHiSeg. The results demonstrate that RevPHiSeg consumes ~30% less memory compared to PHiSeg while achieving very similar segmentation accuracy.
Modelling the Distribution of 3D Brain MRI using a 2D Slice VAE
Jul 09, 2020



Abstract:Probabilistic modelling has been an essential tool in medical image analysis, especially for analyzing brain Magnetic Resonance Images (MRI). Recent deep learning techniques for estimating high-dimensional distributions, in particular Variational Autoencoders (VAEs), opened up new avenues for probabilistic modeling. Modelling of volumetric data has remained a challenge, however, because constraints on available computation and training data make it difficult effectively leverage VAEs, which are well-developed for 2D images. We propose a method to model 3D MR brain volumes distribution by combining a 2D slice VAE with a Gaussian model that captures the relationships between slices. We do so by estimating the sample mean and covariance in the latent space of the 2D model over the slice direction. This combined model lets us sample new coherent stacks of latent variables to decode into slices of a volume. We also introduce a novel evaluation method for generated volumes that quantifies how well their segmentations match those of true brain anatomy. We demonstrate that our proposed model is competitive in generating high quality volumes at high resolutions according to both traditional metrics and our proposed evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge