Xiaoran Chen
GEM: A Generalizable Ego-Vision Multimodal World Model for Fine-Grained Ego-Motion, Object Dynamics, and Scene Composition Control
Dec 15, 2024



Abstract:We present GEM, a Generalizable Ego-vision Multimodal world model that predicts future frames using a reference frame, sparse features, human poses, and ego-trajectories. Hence, our model has precise control over object dynamics, ego-agent motion and human poses. GEM generates paired RGB and depth outputs for richer spatial understanding. We introduce autoregressive noise schedules to enable stable long-horizon generations. Our dataset is comprised of 4000+ hours of multimodal data across domains like autonomous driving, egocentric human activities, and drone flights. Pseudo-labels are used to get depth maps, ego-trajectories, and human poses. We use a comprehensive evaluation framework, including a new Control of Object Manipulation (COM) metric, to assess controllability. Experiments show GEM excels at generating diverse, controllable scenarios and temporal consistency over long generations. Code, models, and datasets are fully open-sourced.
EA-RAS: Towards Efficient and Accurate End-to-End Reconstruction of Anatomical Skeleton
Sep 03, 2024



Abstract:Efficient, accurate and low-cost estimation of human skeletal information is crucial for a range of applications such as biology education and human-computer interaction. However, current simple skeleton models, which are typically based on 2D-3D joint points, fall short in terms of anatomical fidelity, restricting their utility in fields. On the other hand, more complex models while anatomically precise, are hindered by sophisticate multi-stage processing and the need for extra data like skin meshes, making them unsuitable for real-time applications. To this end, we propose the EA-RAS (Towards Efficient and Accurate End-to-End Reconstruction of Anatomical Skeleton), a single-stage, lightweight, and plug-and-play anatomical skeleton estimator that can provide real-time, accurate anatomically realistic skeletons with arbitrary pose using only a single RGB image input. Additionally, EA-RAS estimates the conventional human-mesh model explicitly, which not only enhances the functionality but also leverages the outside skin information by integrating features into the inside skeleton modeling process. In this work, we also develop a progressive training strategy and integrated it with an enhanced optimization process, enabling the network to obtain initial weights using only a small skin dataset and achieve self-supervision in skeleton reconstruction. Besides, we also provide an optional lightweight post-processing optimization strategy to further improve accuracy for scenarios that prioritize precision over real-time processing. The experiments demonstrated that our regression method is over 800 times faster than existing methods, meeting real-time requirements. Additionally, the post-processing optimization strategy provided can enhance reconstruction accuracy by over 50% and achieve a speed increase of more than 7 times.
Modelling the Distribution of 3D Brain MRI using a 2D Slice VAE
Jul 09, 2020



Abstract:Probabilistic modelling has been an essential tool in medical image analysis, especially for analyzing brain Magnetic Resonance Images (MRI). Recent deep learning techniques for estimating high-dimensional distributions, in particular Variational Autoencoders (VAEs), opened up new avenues for probabilistic modeling. Modelling of volumetric data has remained a challenge, however, because constraints on available computation and training data make it difficult effectively leverage VAEs, which are well-developed for 2D images. We propose a method to model 3D MR brain volumes distribution by combining a 2D slice VAE with a Gaussian model that captures the relationships between slices. We do so by estimating the sample mean and covariance in the latent space of the 2D model over the slice direction. This combined model lets us sample new coherent stacks of latent variables to decode into slices of a volume. We also introduce a novel evaluation method for generated volumes that quantifies how well their segmentations match those of true brain anatomy. We demonstrate that our proposed model is competitive in generating high quality volumes at high resolutions according to both traditional metrics and our proposed evaluation.
Unsupervised Lesion Detection via Image Restoration with a Normative Prior
Apr 30, 2020


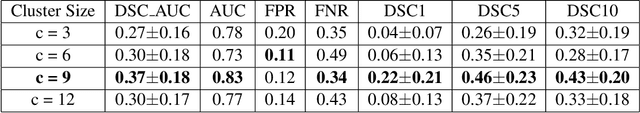
Abstract:Unsupervised lesion detection is a challenging problem that requires accurately estimating normative distributions of healthy anatomy and detecting lesions as outliers without training examples. Recently, this problem has received increased attention from the research community following the advances in unsupervised learning with deep learning. Such advances allow the estimation of high-dimensional distributions, such as normative distributions, with higher accuracy than previous methods.The main approach of the recently proposed methods is to learn a latent-variable model parameterized with networks to approximate the normative distribution using example images showing healthy anatomy, perform prior-projection, i.e. reconstruct the image with lesions using the latent-variable model, and determine lesions based on the differences between the reconstructed and original images. While being promising, the prior-projection step often leads to a large number of false positives. In this work, we approach unsupervised lesion detection as an image restoration problem and propose a probabilistic model that uses a network-based prior as the normative distribution and detect lesions pixel-wise using MAP estimation. The probabilistic model punishes large deviations between restored and original images, reducing false positives in pixel-wise detections. Experiments with gliomas and stroke lesions in brain MRI using publicly available datasets show that the proposed approach outperforms the state-of-the-art unsupervised methods by a substantial margin, +0.13 (AUC), for both glioma and stroke detection. Extensive model analysis confirms the effectiveness of MAP-based image restoration.
Learning Semantic Segmentation from Synthetic Data: A Geometrically Guided Input-Output Adaptation Approach
Jan 13, 2019



Abstract:Recently, increasing attention has been drawn to training semantic segmentation models using synthetic data and computer-generated annotation. However, domain gap remains a major barrier and prevents models learned from synthetic data from generalizing well to real-world applications. In this work, we take the advantage of additional geometric information from synthetic data, a powerful yet largely neglected cue, to bridge the domain gap. Such geometric information can be generated easily from synthetic data, and is proven to be closely coupled with semantic information. With the geometric information, we propose a model to reduce domain shift on two levels: on the input level, we augment the traditional image translation network with the additional geometric information to translate synthetic images into realistic styles; on the output level, we build a task network which simultaneously performs depth estimation and semantic segmentation on the synthetic data. Meanwhile, we encourage the network to preserve correlation between depth and semantics by adversarial training on the output space. We then validate our method on two pairs of synthetic to real dataset: Virtual KITTI to KITTI, and SYNTHIA to Cityscapes, where we achieve a significant performance gain compared to the non-adapt baseline and methods using only semantic label. This demonstrates the usefulness of geometric information from synthetic data for cross-domain semantic segmentation.
Deep Generative Models in the Real-World: An Open Challenge from Medical Imaging
Jun 14, 2018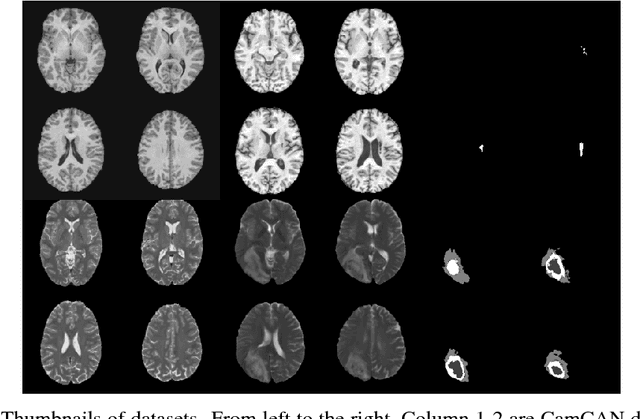
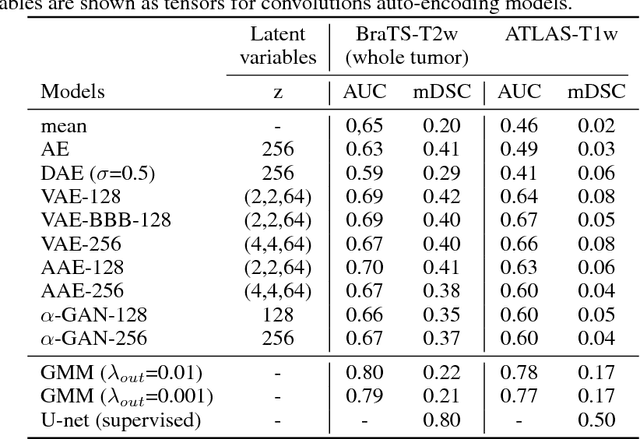
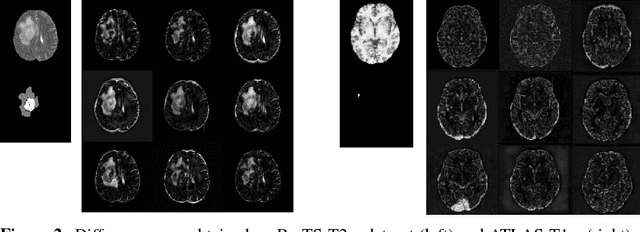
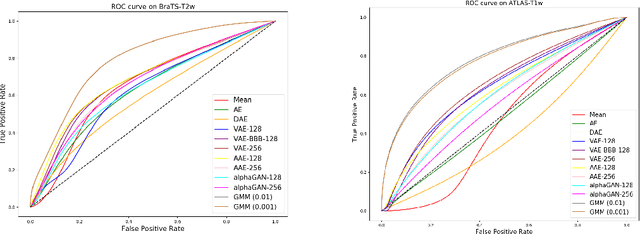
Abstract:Recent advances in deep learning led to novel generative modeling techniques that achieve unprecedented quality in generated samples and performance in learning complex distributions in imaging data. These new models in medical image computing have important applications that form clinically relevant and very challenging unsupervised learning problems. In this paper, we explore the feasibility of using state-of-the-art auto-encoder-based deep generative models, such as variational and adversarial auto-encoders, for one such task: abnormality detection in medical imaging. We utilize typical, publicly available datasets with brain scans from healthy subjects and patients with stroke lesions and brain tumors. We use the data from healthy subjects to train different auto-encoder based models to learn the distribution of healthy images and detect pathologies as outliers. Models that can better learn the data distribution should be able to detect outliers more accurately. We evaluate the detection performance of deep generative models and compare them with non-deep learning based approaches to provide a benchmark of the current state of research. We conclude that abnormality detection is a challenging task for deep generative models and large room exists for improvement. In order to facilitate further research, we aim to provide carefully pre-processed imaging data available to the research community.
Unsupervised Detection of Lesions in Brain MRI using constrained adversarial auto-encoders
Jun 13, 2018

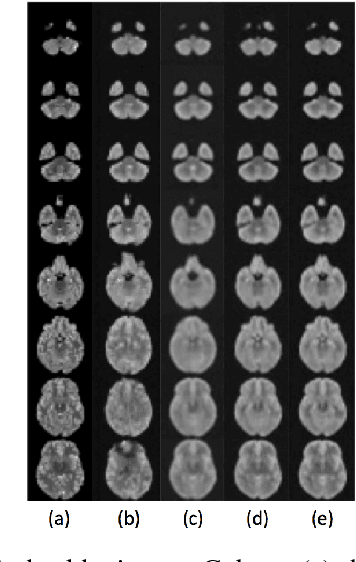
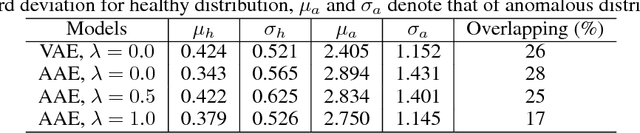
Abstract:Lesion detection in brain Magnetic Resonance Images (MRI) remains a challenging task. State-of-the-art approaches are mostly based on supervised learning making use of large annotated datasets. Human beings, on the other hand, even non-experts, can detect most abnormal lesions after seeing a handful of healthy brain images. Replicating this capability of using prior information on the appearance of healthy brain structure to detect lesions can help computers achieve human level abnormality detection, specifically reducing the need for numerous labeled examples and bettering generalization of previously unseen lesions. To this end, we study detection of lesion regions in an unsupervised manner by learning data distribution of brain MRI of healthy subjects using auto-encoder based methods. We hypothesize that one of the main limitations of the current models is the lack of consistency in latent representation. We propose a simple yet effective constraint that helps mapping of an image bearing lesion close to its corresponding healthy image in the latent space. We use the Human Connectome Project dataset to learn distribution of healthy-appearing brain MRI and report improved detection, in terms of AUC, of the lesions in the BRATS challenge dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge