Gustav Bredell
Explicit and data-Efficient Encoding via Gradient Flow
Dec 01, 2024Abstract:The autoencoder model typically uses an encoder to map data to a lower dimensional latent space and a decoder to reconstruct it. However, relying on an encoder for inversion can lead to suboptimal representations, particularly limiting in physical sciences where precision is key. We introduce a decoder-only method using gradient flow to directly encode data into the latent space, defined by ordinary differential equations (ODEs). This approach eliminates the need for approximate encoder inversion. We train the decoder via the adjoint method and show that costly integrals can be avoided with minimal accuracy loss. Additionally, we propose a $2^{nd}$ order ODE variant, approximating Nesterov's accelerated gradient descent for faster convergence. To handle stiff ODEs, we use an adaptive solver that prioritizes loss minimization, improving robustness. Compared to traditional autoencoders, our method demonstrates explicit encoding and superior data efficiency, which is crucial for data-scarce scenarios in the physical sciences. Furthermore, this work paves the way for integrating machine learning into scientific workflows, where precise and efficient encoding is critical. \footnote{The code for this work is available at \url{https://github.com/k-flouris/gfe}.}
Aggregation Model Hyperparameters Matter in Digital Pathology
Nov 29, 2023Abstract:Digital pathology has significantly advanced disease detection and pathologist efficiency through the analysis of gigapixel whole-slide images (WSI). In this process, WSIs are first divided into patches, for which a feature extractor model is applied to obtain feature vectors, which are subsequently processed by an aggregation model to predict the respective WSI label. With the rapid evolution of representation learning, numerous new feature extractor models, often termed foundational models, have emerged. Traditional evaluation methods, however, rely on fixed aggregation model hyperparameters, a framework we identify as potentially biasing the results. Our study uncovers a co-dependence between feature extractor models and aggregation model hyperparameters, indicating that performance comparability can be skewed based on the chosen hyperparameters. By accounting for this co-dependency, we find that the performance of many current feature extractor models is notably similar. We support this insight by evaluating seven feature extractor models across three different datasets with 162 different aggregation model configurations. This comprehensive approach provides a more nuanced understanding of the relationship between feature extractors and aggregation models, leading to a fairer and more accurate assessment of feature extractor models in digital pathology.
Explicitly Minimizing the Blur Error of Variational Autoencoders
Apr 12, 2023



Abstract:Variational autoencoders (VAEs) are powerful generative modelling methods, however they suffer from blurry generated samples and reconstructions compared to the images they have been trained on. Significant research effort has been spent to increase the generative capabilities by creating more flexible models but often flexibility comes at the cost of higher complexity and computational cost. Several works have focused on altering the reconstruction term of the evidence lower bound (ELBO), however, often at the expense of losing the mathematical link to maximizing the likelihood of the samples under the modeled distribution. Here we propose a new formulation of the reconstruction term for the VAE that specifically penalizes the generation of blurry images while at the same time still maximizing the ELBO under the modeled distribution. We show the potential of the proposed loss on three different data sets, where it outperforms several recently proposed reconstruction losses for VAEs.
Unsupervised Superpixel Generation using Edge-Sparse Embedding
Nov 29, 2022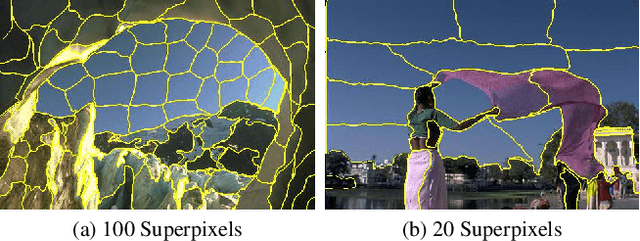
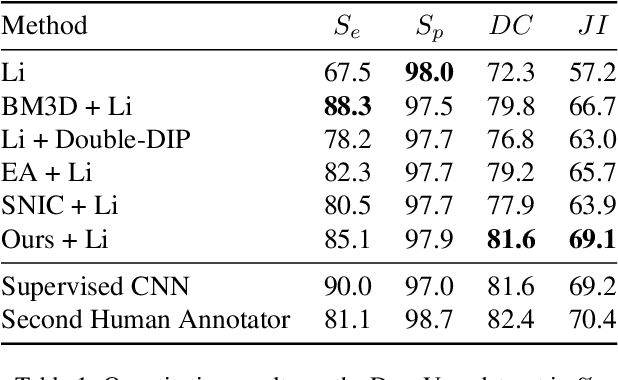
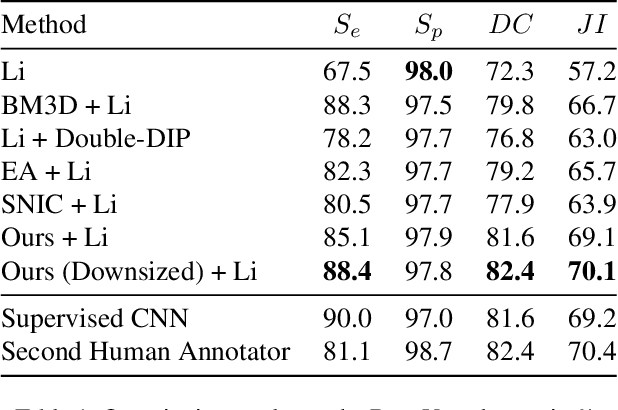
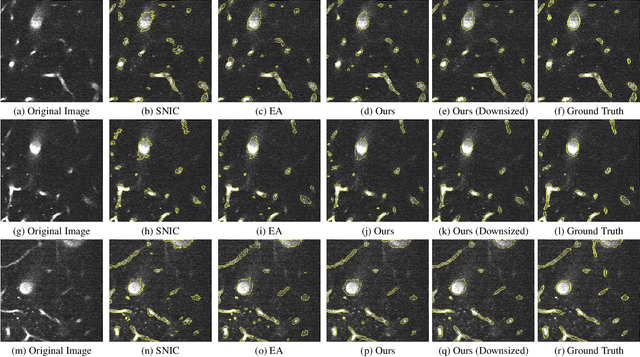
Abstract:Partitioning an image into superpixels based on the similarity of pixels with respect to features such as colour or spatial location can significantly reduce data complexity and improve subsequent image processing tasks. Initial algorithms for unsupervised superpixel generation solely relied on local cues without prioritizing significant edges over arbitrary ones. On the other hand, more recent methods based on unsupervised deep learning either fail to properly address the trade-off between superpixel edge adherence and compactness or lack control over the generated number of superpixels. By using random images with strong spatial correlation as input, \ie, blurred noise images, in a non-convolutional image decoder we can reduce the expected number of contrasts and enforce smooth, connected edges in the reconstructed image. We generate edge-sparse pixel embeddings by encoding additional spatial information into the piece-wise smooth activation maps from the decoder's last hidden layer and use a standard clustering algorithm to extract high quality superpixels. Our proposed method reaches state-of-the-art performance on the BSDS500, PASCAL-Context and a microscopy dataset.
Wiener Guided DIP for Unsupervised Blind Image Deconvolution
Dec 19, 2021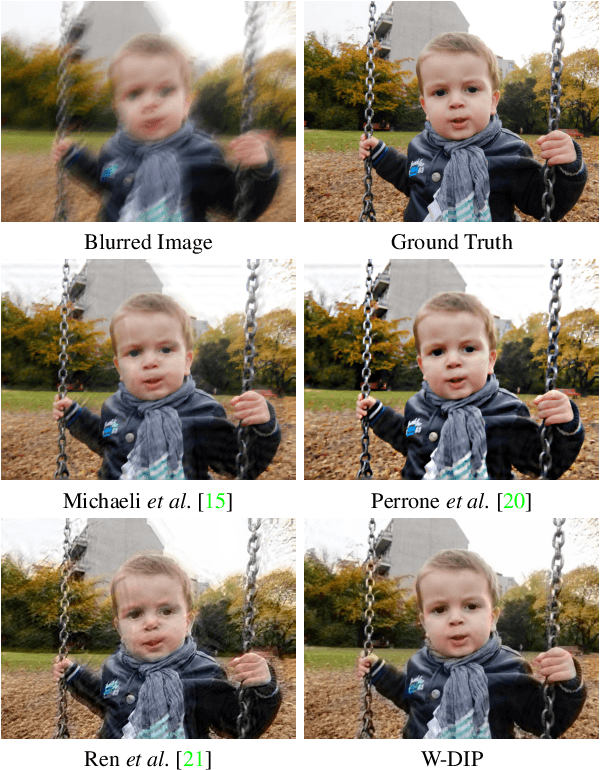
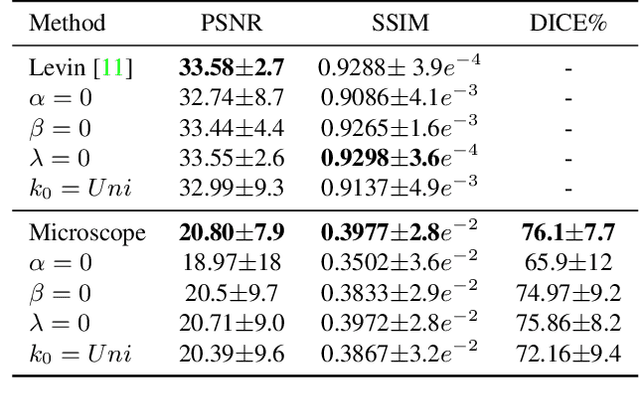
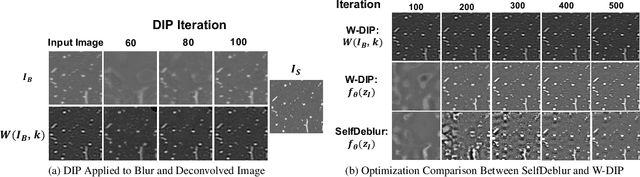
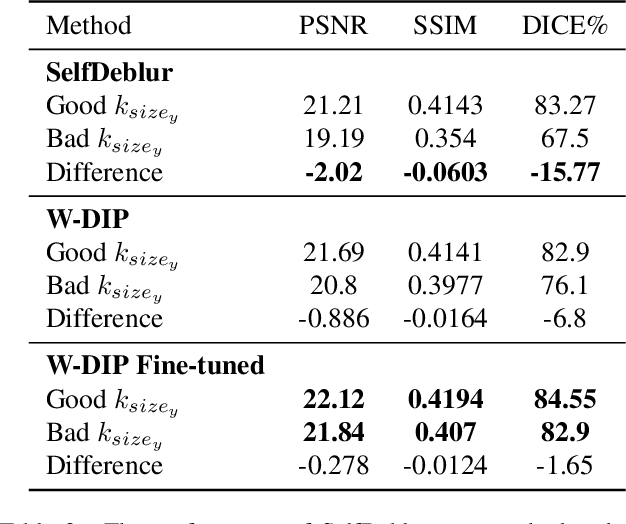
Abstract:Blind deconvolution is an ill-posed problem arising in various fields ranging from microscopy to astronomy. The ill-posed nature of the problem requires adequate priors to arrive to a desirable solution. Recently, it has been shown that deep learning architectures can serve as an image generation prior during unsupervised blind deconvolution optimization, however often exhibiting a performance fluctuation even on a single image. We propose to use Wiener-deconvolution to guide the image generator during optimization by providing it a sharpened version of the blurry image using an auxiliary kernel estimate starting from a Gaussian. We observe that the high-frequency artifacts of deconvolution are reproduced with a delay compared to low-frequency features. In addition, the image generator reproduces low-frequency features of the deconvolved image faster than that of a blurry image. We embed the computational process in a constrained optimization framework and show that the proposed method yields higher stability and performance across multiple datasets. In addition, we provide the code.
ISNAS-DIP: Image-Specific Neural Architecture Search for Deep Image Prior
Nov 27, 2021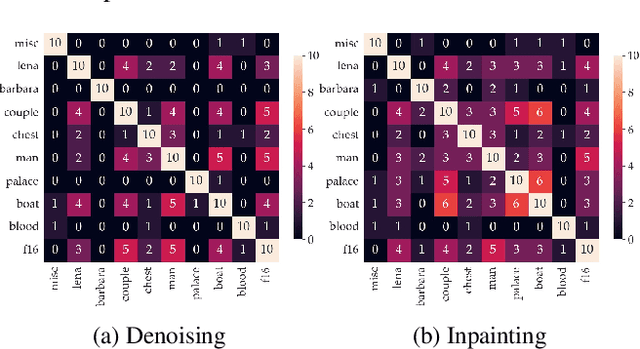
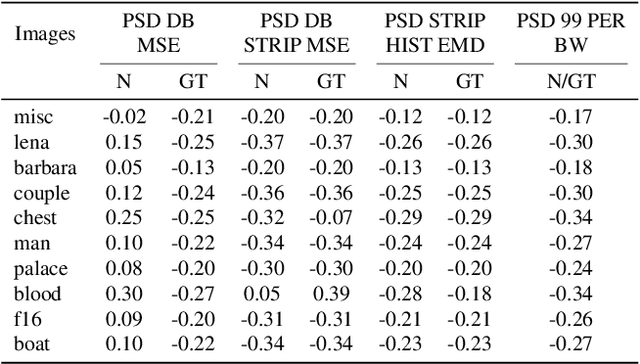
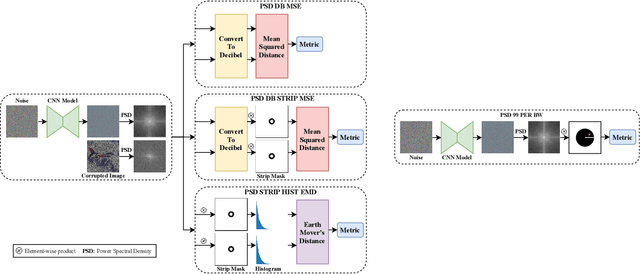
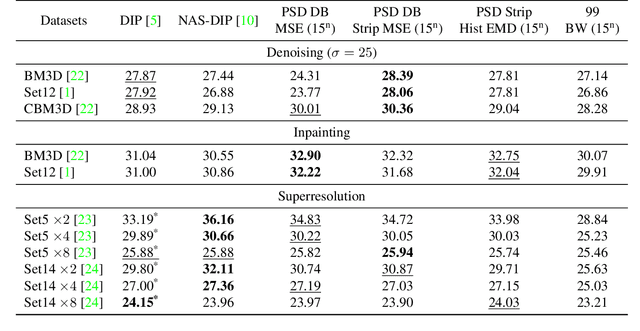
Abstract:Recent works show that convolutional neural network (CNN) architectures have a spectral bias towards lower frequencies, which has been leveraged for various image restoration tasks in the Deep Image Prior (DIP) framework. The benefit of the inductive bias the network imposes in the DIP framework depends on the architecture. Therefore, researchers have studied how to automate the search to determine the best-performing model. However, common neural architecture search (NAS) techniques are resource and time-intensive. Moreover, best-performing models are determined for a whole dataset of images instead of for each image independently, which would be prohibitively expensive. In this work, we first show that optimal neural architectures in the DIP framework are image-dependent. Leveraging this insight, we then propose an image-specific NAS strategy for the DIP framework that requires substantially less training than typical NAS approaches, effectively enabling image-specific NAS. For a given image, noise is fed to a large set of untrained CNNs, and their outputs' power spectral densities (PSD) are compared to that of the corrupted image using various metrics. Based on this, a small cohort of image-specific architectures is chosen and trained to reconstruct the corrupted image. Among this cohort, the model whose reconstruction is closest to the average of the reconstructed images is chosen as the final model. We justify the proposed strategy's effectiveness by (1) demonstrating its performance on a NAS Dataset for DIP that includes 500+ models from a particular search space (2) conducting extensive experiments on image denoising, inpainting, and super-resolution tasks. Our experiments show that image-specific metrics can reduce the search space to a small cohort of models, of which the best model outperforms current NAS approaches for image restoration.
Quality-Aware Memory Network for Interactive Volumetric Image Segmentation
Jul 05, 2021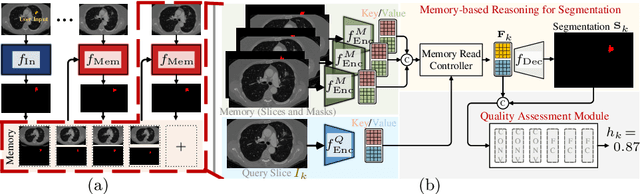

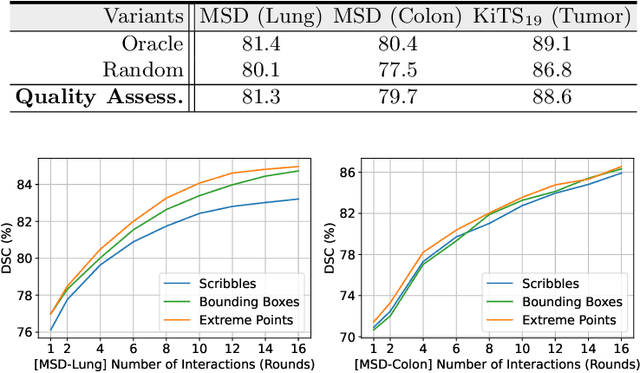
Abstract:Despite recent progress of automatic medical image segmentation techniques, fully automatic results usually fail to meet the clinical use and typically require further refinement. In this work, we propose a quality-aware memory network for interactive segmentation of 3D medical images. Provided by user guidance on an arbitrary slice, an interaction network is firstly employed to obtain an initial 2D segmentation. The quality-aware memory network subsequently propagates the initial segmentation estimation bidirectionally over the entire volume. Subsequent refinement based on additional user guidance on other slices can be incorporated in the same manner. To further facilitate interactive segmentation, a quality assessment module is introduced to suggest the next slice to segment based on the current segmentation quality of each slice. The proposed network has two appealing characteristics: 1) The memory-augmented network offers the ability to quickly encode past segmentation information, which will be retrieved for the segmentation of other slices; 2) The quality assessment module enables the model to directly estimate the qualities of segmentation predictions, which allows an active learning paradigm where users preferentially label the lowest-quality slice for multi-round refinement. The proposed network leads to a robust interactive segmentation engine, which can generalize well to various types of user annotations (e.g., scribbles, boxes). Experimental results on various medical datasets demonstrate the superiority of our approach in comparison with existing techniques.
Gradient flow encoding with distance optimization adaptive step size
May 11, 2021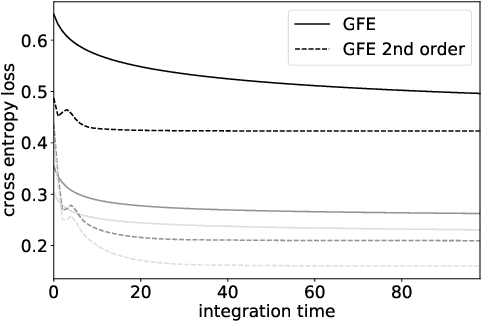

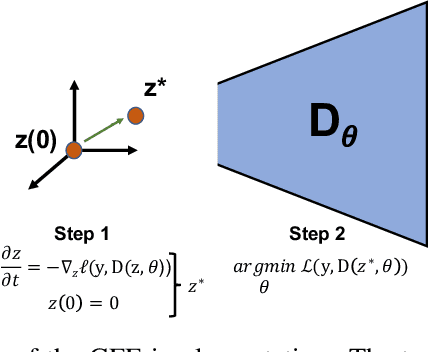
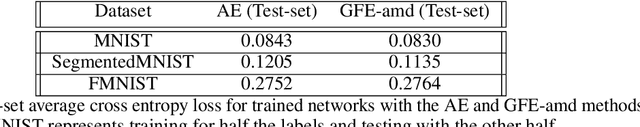
Abstract:The autoencoder model uses an encoder to map data samples to a lower dimensional latent space and then a decoder to map the latent space representations back to the data space. Implicitly, it relies on the encoder to approximate the inverse of the decoder network, so that samples can be mapped to and back from the latent space faithfully. This approximation may lead to sub-optimal latent space representations. In this work, we investigate a decoder-only method that uses gradient flow to encode data samples in the latent space. The gradient flow is defined based on a given decoder and aims to find the optimal latent space representation for any given sample through optimisation, eliminating the need of an approximate inversion through an encoder. Implementing gradient flow through ordinary differential equations (ODE), we leverage the adjoint method to train a given decoder. We further show empirically that the costly integrals in the adjoint method may not be entirely necessary. Additionally, we propose a $2^{nd}$ order ODE variant to the method, which approximates Nesterov's accelerated gradient descent, with faster convergence per iteration. Commonly used ODE solvers can be quite sensitive to the integration step-size depending on the stiffness of the ODE. To overcome the sensitivity for gradient flow encoding, we use an adaptive solver that prioritises minimising loss at each integration step. We assess the proposed method in comparison to the autoencoding model. In our experiments, GFE showed a much higher data-efficiency than the autoencoding model, which can be crucial for data scarce applications.
Iterative Interaction Training for Segmentation Editing Networks
Jul 23, 2018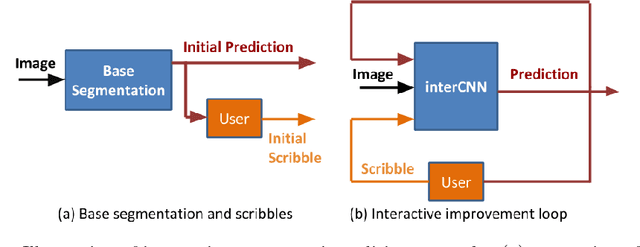
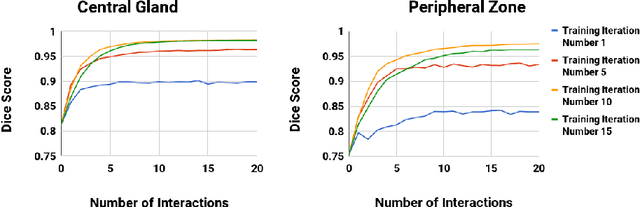
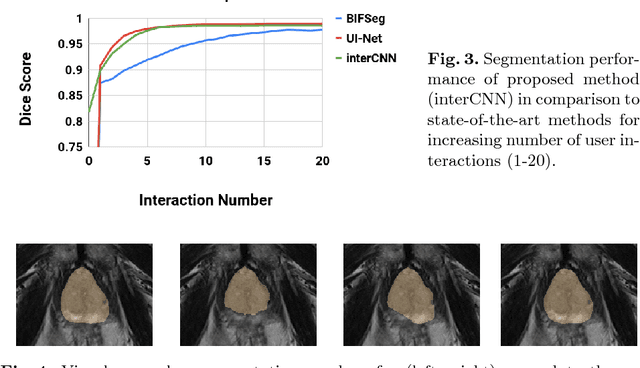
Abstract:Automatic segmentation has great potential to facilitate morphological measurements while simultaneously increasing efficiency. Nevertheless often users want to edit the segmentation to their own needs and will need different tools for this. There has been methods developed to edit segmentations of automatic methods based on the user input, primarily for binary segmentations. Here however, we present an unique training strategy for convolutional neural networks (CNNs) trained on top of an automatic method to enable interactive segmentation editing that is not limited to binary segmentation. By utilizing a robot-user during training, we closely mimic realistic use cases to achieve optimal editing performance. In addition, we show that an increase of the iterative interactions during the training process up to ten improves the segmentation editing performance substantially. Furthermore, we compare our segmentation editing CNN (interCNN) to state-of-the-art interactive segmentation algorithms and show a superior or on par performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge