Samaneh Abbasi-Sureshjani
Integrating multiscale topology in digital pathology with pyramidal graph convolutional networks
Mar 22, 2024Abstract:Graph convolutional networks (GCNs) have emerged as a powerful alternative to multiple instance learning with convolutional neural networks in digital pathology, offering superior handling of structural information across various spatial ranges - a crucial aspect of learning from gigapixel H&E-stained whole slide images (WSI). However, graph message-passing algorithms often suffer from oversmoothing when aggregating a large neighborhood. Hence, effective modeling of multi-range interactions relies on the careful construction of the graph. Our proposed multi-scale GCN (MS-GCN) tackles this issue by leveraging information across multiple magnification levels in WSIs. MS-GCN enables the simultaneous modeling of long-range structural dependencies at lower magnifications and high-resolution cellular details at higher magnifications, akin to analysis pipelines usually conducted by pathologists. The architecture's unique configuration allows for the concurrent modeling of structural patterns at lower magnifications and detailed cellular features at higher ones, while also quantifying the contribution of each magnification level to the prediction. Through testing on different datasets, MS-GCN demonstrates superior performance over existing single-magnification GCN methods. The enhancement in performance and interpretability afforded by our method holds promise for advancing computational pathology models, especially in tasks requiring extensive spatial context.
Aggregation Model Hyperparameters Matter in Digital Pathology
Nov 29, 2023Abstract:Digital pathology has significantly advanced disease detection and pathologist efficiency through the analysis of gigapixel whole-slide images (WSI). In this process, WSIs are first divided into patches, for which a feature extractor model is applied to obtain feature vectors, which are subsequently processed by an aggregation model to predict the respective WSI label. With the rapid evolution of representation learning, numerous new feature extractor models, often termed foundational models, have emerged. Traditional evaluation methods, however, rely on fixed aggregation model hyperparameters, a framework we identify as potentially biasing the results. Our study uncovers a co-dependence between feature extractor models and aggregation model hyperparameters, indicating that performance comparability can be skewed based on the chosen hyperparameters. By accounting for this co-dependency, we find that the performance of many current feature extractor models is notably similar. We support this insight by evaluating seven feature extractor models across three different datasets with 162 different aggregation model configurations. This comprehensive approach provides a more nuanced understanding of the relationship between feature extractors and aggregation models, leading to a fairer and more accurate assessment of feature extractor models in digital pathology.
A comparative study between vision transformers and CNNs in digital pathology
Jun 01, 2022



Abstract:Recently, vision transformers were shown to be capable of outperforming convolutional neural networks when pretrained on sufficient amounts of data. In comparison to convolutional neural networks, vision transformers have a weaker inductive bias and therefore allow a more flexible feature detection. Due to their promising feature detection, this work explores vision transformers for tumor detection in digital pathology whole slide images in four tissue types, and for tissue type identification. We compared the patch-wise classification performance of the vision transformer DeiT-Tiny to the state-of-the-art convolutional neural network ResNet18. Due to the sparse availability of annotated whole slide images, we further compared both models pretrained on large amounts of unlabeled whole-slide images using state-of-the-art self-supervised approaches. The results show that the vision transformer performed slightly better than the ResNet18 for three of four tissue types for tumor detection while the ResNet18 performed slightly better for the remaining tasks. The aggregated predictions of both models on slide level were correlated, indicating that the models captured similar imaging features. All together, the vision transformer models performed on par with the ResNet18 while requiring more effort to train. In order to surpass the performance of convolutional neural networks, vision transformers might require more challenging tasks to benefit from their weak inductive bias.
XCAT-GAN for Synthesizing 3D Consistent Labeled Cardiac MR Images on Anatomically Variable XCAT Phantoms
Jul 31, 2020



Abstract:Generative adversarial networks (GANs) have provided promising data enrichment solutions by synthesizing high-fidelity images. However, generating large sets of labeled images with new anatomical variations remains unexplored. We propose a novel method for synthesizing cardiac magnetic resonance (CMR) images on a population of virtual subjects with a large anatomical variation, introduced using the 4D eXtended Cardiac and Torso (XCAT) computerized human phantom. We investigate two conditional image synthesis approaches grounded on a semantically-consistent mask-guided image generation technique: 4-class and 8-class XCAT-GANs. The 4-class technique relies on only the annotations of the heart; while the 8-class technique employs a predicted multi-tissue label map of the heart-surrounding organs and provides better guidance for our conditional image synthesis. For both techniques, we train our conditional XCAT-GAN with real images paired with corresponding labels and subsequently at the inference time, we substitute the labels with the XCAT derived ones. Therefore, the trained network accurately transfers the tissue-specific textures to the new label maps. By creating 33 virtual subjects of synthetic CMR images at the end-diastolic and end-systolic phases, we evaluate the usefulness of such data in the downstream cardiac cavity segmentation task under different augmentation strategies. Results demonstrate that even with only 20% of real images (40 volumes) seen during training, segmentation performance is retained with the addition of synthetic CMR images. Moreover, the improvement in utilizing synthetic images for augmenting the real data is evident through the reduction of Hausdorff distance up to 28% and an increase in the Dice score up to 5%, indicating a higher similarity to the ground truth in all dimensions.
Risk of Training Diagnostic Algorithms on Data with Demographic Bias
Jun 17, 2020


Abstract:One of the critical challenges in machine learning applications is to have fair predictions. There are numerous recent examples in various domains that convincingly show that algorithms trained with biased datasets can easily lead to erroneous or discriminatory conclusions. This is even more crucial in clinical applications where the predictive algorithms are designed mainly based on a limited or given set of medical images and demographic variables such as age, sex and race are not taken into account. In this work, we conduct a survey of the MICCAI 2018 proceedings to investigate the common practice in medical image analysis applications. Surprisingly, we found that papers focusing on diagnosis rarely describe the demographics of the datasets used, and the diagnosis is purely based on images. In order to highlight the importance of considering the demographics in diagnosis tasks, we used a publicly available dataset of skin lesions. We then demonstrate that a classifier with an overall area under the curve (AUC) of 0.83 has variable performance between 0.76 and 0.91 on subgroups based on age and sex, even though the training set was relatively balanced. Moreover, we show that it is possible to learn unbiased features by explicitly using demographic variables in an adversarial training setup, which leads to balanced scores per subgroups. Finally, we discuss the implications of these results and provide recommendations for further research.
4D Semantic Cardiac Magnetic Resonance Image Synthesis on XCAT Anatomical Model
Feb 17, 2020



Abstract:We propose a hybrid controllable image generation method to synthesize anatomically meaningful 3D+t labeled Cardiac Magnetic Resonance (CMR) images. Our hybrid method takes the mechanistic 4D eXtended CArdiac Torso (XCAT) heart model as the anatomical ground truth and synthesizes CMR images via a data-driven Generative Adversarial Network (GAN). We employ the state-of-the-art SPatially Adaptive De-normalization (SPADE) technique for conditional image synthesis to preserve the semantic spatial information of ground truth anatomy. Using the parameterized motion model of the XCAT heart, we generate labels for 25 time frames of the heart for one cardiac cycle at 18 locations for the short axis view. Subsequently, realistic images are generated from these labels, with modality-specific features that are learned from real CMR image data. We demonstrate that style transfer from another cardiac image can be accomplished by using a style encoder network. Due to the flexibility of XCAT in creating new heart models, this approach can result in a realistic virtual population to address different challenges the medical image analysis research community is facing such as expensive data collection. Our proposed method has a great potential to synthesize 4D controllable CMR images with annotations and adaptable styles to be used in various supervised multi-site, multi-vendor applications in medical image analysis.
Curvature Integration in a 5D Kernel for Extracting Vessel Connections in Retinal Images
Jun 26, 2017
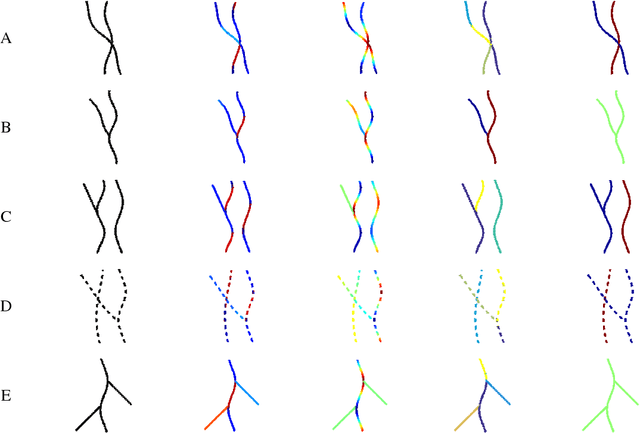


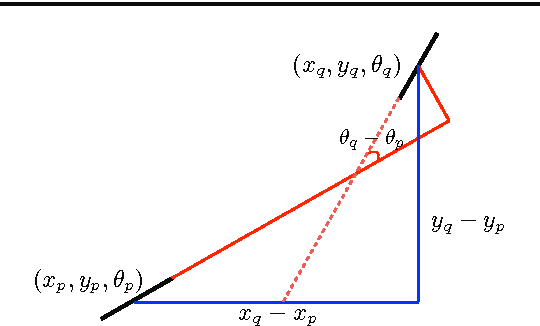
Abstract:Tree-like structures such as retinal images are widely studied in computer-aided diagnosis systems for large-scale screening programs. Despite several segmentation and tracking methods proposed in the literature, there still exist several limitations specifically when two or more curvilinear structures cross or bifurcate, or in the presence of interrupted lines or highly curved blood vessels. In this paper, we propose a novel approach based on multi-orientation scores augmented with a contextual affinity matrix, which both are inspired by the geometry of the primary visual cortex (V1) and their contextual connections. The connectivity is described with a five-dimensional kernel obtained as the fundamental solution of the Fokker-Planck equation modelling the cortical connectivity in the lifted space of positions, orientations, curvatures and intensity. It is further used in a self-tuning spectral clustering step to identify the main perceptual units in the stimuli. The proposed method has been validated on several easy and challenging structures in a set of artificial images and actual retinal patches. Supported by quantitative and qualitative results, the method is capable of overcoming the limitations of current state-of-the-art techniques.
Retrieving challenging vessel connections in retinal images by line co-occurrence statistics
Oct 20, 2016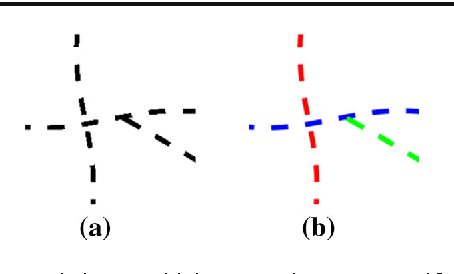
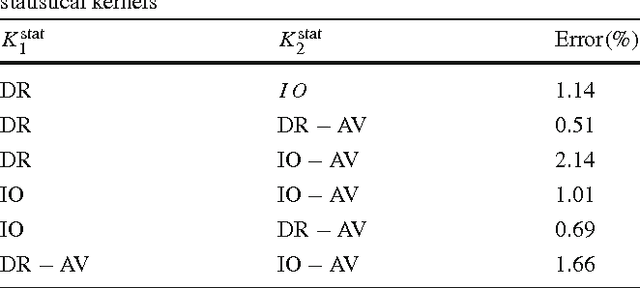
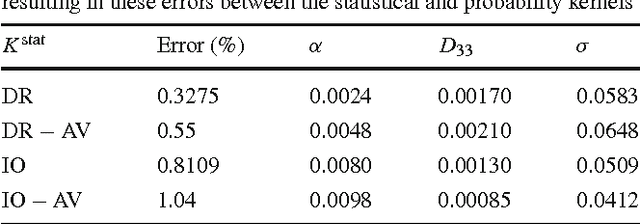
Abstract:Natural images contain often curvilinear structures, which might be disconnected, or partly occluded. Recovering the missing connection of disconnected structures is an open issue and needs appropriate geometric reasoning. We propose to find line co-occurrence statistics from the centerlines of blood vessels in retinal images and show its remarkable similarity to a well-known probabilistic model for the connectivity pattern in the primary visual cortex. Furthermore, the probabilistic model is trained from the data via statistics and used for automated grouping of interrupted vessels in a spectral clustering based approach. Several challenging image patches are investigated around junction points, where successful results indicate the perfect match of the trained model to the profiles of blood vessels in retinal images. Also, comparisons among several statistical models obtained from different datasets reveals their high similarity i.e., they are independent of the dataset. On top of that, the best approximation of the statistical model with the symmetrized extension of the probabilistic model on the projective line bundle is found with a least square error smaller than 2%. Apparently, the direction process on the projective line bundle is a good continuation model for vessels in retinal images.
Analysis of Vessel Connectivities in Retinal Images by Cortically Inspired Spectral Clustering
May 23, 2016

Abstract:Retinal images provide early signs of diabetic retinopathy, glaucoma, and hypertension. These signs can be investigated based on microaneurysms or smaller vessels. The diagnostic biomarkers are the change of vessel widths and angles especially at junctions, which are investigated using the vessel segmentation or tracking. Vessel paths may also be interrupted; crossings and bifurcations may be disconnected. This paper addresses a novel contextual method based on the geometry of the primary visual cortex (V1) to study these difficulties. We have analyzed the specific problems at junctions with a connectivity kernel obtained as the fundamental solution of the Fokker-Planck equation, which is usually used to represent the geometrical structure of multi-orientation cortical connectivity. Using the spectral clustering on a large local affinity matrix constructed by both the connectivity kernel and the feature of intensity, the vessels are identified successfully in a hierarchical topology each representing an individual perceptual unit.
* submitted to and accepted by JMIV
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge