Sina Amirrajab
Large-scale modality-invariant foundation models for brain MRI analysis: Application to lesion segmentation
Nov 14, 2025Abstract:The field of computer vision is undergoing a paradigm shift toward large-scale foundation model pre-training via self-supervised learning (SSL). Leveraging large volumes of unlabeled brain MRI data, such models can learn anatomical priors that improve few-shot performance in diverse neuroimaging tasks. However, most SSL frameworks are tailored to natural images, and their adaptation to capture multi-modal MRI information remains underexplored. This work proposes a modality-invariant representation learning setup and evaluates its effectiveness in stroke and epilepsy lesion segmentation, following large-scale pre-training. Experimental results suggest that despite successful cross-modality alignment, lesion segmentation primarily benefits from preserving fine-grained modality-specific features. Model checkpoints and code are made publicly available.
Radiology Report Conditional 3D CT Generation with Multi Encoder Latent diffusion Model
Sep 18, 2025



Abstract:Text to image latent diffusion models have recently advanced medical image synthesis, but applications to 3D CT generation remain limited. Existing approaches rely on simplified prompts, neglecting the rich semantic detail in full radiology reports, which reduces text image alignment and clinical fidelity. We propose Report2CT, a radiology report conditional latent diffusion framework for synthesizing 3D chest CT volumes directly from free text radiology reports, incorporating both findings and impression sections using multiple text encoder. Report2CT integrates three pretrained medical text encoders (BiomedVLP CXR BERT, MedEmbed, and ClinicalBERT) to capture nuanced clinical context. Radiology reports and voxel spacing information condition a 3D latent diffusion model trained on 20000 CT volumes from the CT RATE dataset. Model performance was evaluated using Frechet Inception Distance (FID) for real synthetic distributional similarity and CLIP based metrics for semantic alignment, with additional qualitative and quantitative comparisons against GenerateCT model. Report2CT generated anatomically consistent CT volumes with excellent visual quality and text image alignment. Multi encoder conditioning improved CLIP scores, indicating stronger preservation of fine grained clinical details in the free text radiology reports. Classifier free guidance further enhanced alignment with only a minor trade off in FID. We ranked first in the VLM3D Challenge at MICCAI 2025 on Text Conditional CT Generation and achieved state of the art performance across all evaluation metrics. By leveraging complete radiology reports and multi encoder text conditioning, Report2CT advances 3D CT synthesis, producing clinically faithful and high quality synthetic data.
A Foundation Model Framework for Multi-View MRI Classification of Extramural Vascular Invasion and Mesorectal Fascia Invasion in Rectal Cancer
May 23, 2025Abstract:Background: Accurate MRI-based identification of extramural vascular invasion (EVI) and mesorectal fascia invasion (MFI) is pivotal for risk-stratified management of rectal cancer, yet visual assessment is subjective and vulnerable to inter-institutional variability. Purpose: To develop and externally evaluate a multicenter, foundation-model-driven framework that automatically classifies EVI and MFI on axial and sagittal T2-weighted MRI. Methods: This retrospective study used 331 pre-treatment rectal cancer MRI examinations from three European hospitals. After TotalSegmentator-guided rectal patch extraction, a self-supervised frequency-domain harmonization pipeline was trained to minimize scanner-related contrast shifts. Four classifiers were compared: ResNet50, SeResNet, the universal biomedical pretrained transformer (UMedPT) with a lightweight MLP head, and a logistic-regression variant using frozen UMedPT features (UMedPT_LR). Results: UMedPT_LR achieved the best EVI detection when axial and sagittal features were fused (AUC = 0.82; sensitivity = 0.75; F1 score = 0.73), surpassing the Chaimeleon Grand-Challenge winner (AUC = 0.74). The highest MFI performance was attained by UMedPT on axial harmonized images (AUC = 0.77), surpassing the Chaimeleon Grand-Challenge winner (AUC = 0.75). Frequency-domain harmonization improved MFI classification but variably affected EVI performance. Conventional CNNs (ResNet50, SeResNet) underperformed, especially in F1 score and balanced accuracy. Conclusion: These findings demonstrate that combining foundation model features, harmonization, and multi-view fusion significantly enhances diagnostic performance in rectal MRI.
Explainable Anatomy-Guided AI for Prostate MRI: Foundation Models and In Silico Clinical Trials for Virtual Biopsy-based Risk Assessment
May 23, 2025Abstract:We present a fully automated, anatomically guided deep learning pipeline for prostate cancer (PCa) risk stratification using routine MRI. The pipeline integrates three key components: an nnU-Net module for segmenting the prostate gland and its zones on axial T2-weighted MRI; a classification module based on the UMedPT Swin Transformer foundation model, fine-tuned on 3D patches with optional anatomical priors and clinical data; and a VAE-GAN framework for generating counterfactual heatmaps that localize decision-driving image regions. The system was developed using 1,500 PI-CAI cases for segmentation and 617 biparametric MRIs with metadata from the CHAIMELEON challenge for classification (split into 70% training, 10% validation, and 20% testing). Segmentation achieved mean Dice scores of 0.95 (gland), 0.94 (peripheral zone), and 0.92 (transition zone). Incorporating gland priors improved AUC from 0.69 to 0.72, with a three-scale ensemble achieving top performance (AUC = 0.79, composite score = 0.76), outperforming the 2024 CHAIMELEON challenge winners. Counterfactual heatmaps reliably highlighted lesions within segmented regions, enhancing model interpretability. In a prospective multi-center in-silico trial with 20 clinicians, AI assistance increased diagnostic accuracy from 0.72 to 0.77 and Cohen's kappa from 0.43 to 0.53, while reducing review time per case by 40%. These results demonstrate that anatomy-aware foundation models with counterfactual explainability can enable accurate, interpretable, and efficient PCa risk assessment, supporting their potential use as virtual biopsies in clinical practice.
Enhancing Reconstruction-Based Out-of-Distribution Detection in Brain MRI with Model and Metric Ensembles
Dec 23, 2024Abstract:Out-of-distribution (OOD) detection is crucial for safely deploying automated medical image analysis systems, as abnormal patterns in images could hamper their performance. However, OOD detection in medical imaging remains an open challenge, and we address three gaps: the underexplored potential of a simple OOD detection model, the lack of optimization of deep learning strategies specifically for OOD detection, and the selection of appropriate reconstruction metrics. In this study, we investigated the effectiveness of a reconstruction-based autoencoder for unsupervised detection of synthetic artifacts in brain MRI. We evaluated the general reconstruction capability of the model, analyzed the impact of the selected training epoch and reconstruction metrics, assessed the potential of model and/or metric ensembles, and tested the model on a dataset containing a diverse range of artifacts. Among the metrics assessed, the contrast component of SSIM and LPIPS consistently outperformed others in detecting homogeneous circular anomalies. By combining two well-converged models and using LPIPS and contrast as reconstruction metrics, we achieved a pixel-level area under the Precision-Recall curve of 0.66. Furthermore, with the more realistic OOD dataset, we observed that the detection performance varied between artifact types; local artifacts were more difficult to detect, while global artifacts showed better detection results. These findings underscore the importance of carefully selecting metrics and model configurations, and highlight the need for tailored approaches, as standard deep learning approaches do not always align with the unique needs of OOD detection.
WAND: Wavelet Analysis-based Neural Decomposition of MRS Signals for Artifact Removal
Oct 14, 2024Abstract:Accurate quantification of metabolites in magnetic resonance spectroscopy (MRS) is challenged by low signal-to-noise ratio (SNR), overlapping metabolites, and various artifacts. Particularly, unknown and unparameterized baseline effects obscure the quantification of low-concentration metabolites, limiting MRS reliability. This paper introduces wavelet analysis-based neural decomposition (WAND), a novel data-driven method designed to decompose MRS signals into their constituent components: metabolite-specific signals, baseline, and artifacts. WAND takes advantage of the enhanced separability of these components within the wavelet domain. The method employs a neural network, specifically a U-Net architecture, trained to predict masks for wavelet coefficients obtained through the continuous wavelet transform. These masks effectively isolate desired signal components in the wavelet domain, which are then inverse-transformed to obtain separated signals. Notably, an artifact mask is created by inverting the sum of all known signal masks, enabling WAND to capture and remove even unpredictable artifacts. The effectiveness of WAND in achieving accurate decomposition is demonstrated through numerical evaluations using simulated spectra. Furthermore, WAND's artifact removal capabilities significantly enhance the quantification accuracy of linear combination model fitting. The method's robustness is further validated using data from the 2016 MRS Fitting Challenge and in-vivo experiments.
Generative AI for Synthetic Data Across Multiple Medical Modalities: A Systematic Review of Recent Developments and Challenges
Jul 02, 2024


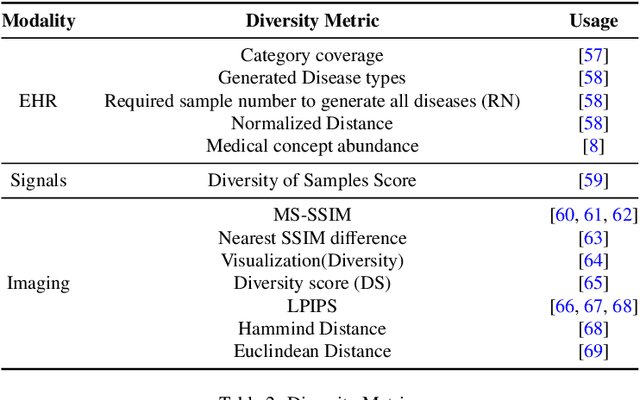
Abstract:This paper presents a comprehensive systematic review of generative models (GANs, VAEs, DMs, and LLMs) used to synthesize various medical data types, including imaging (dermoscopic, mammographic, ultrasound, CT, MRI, and X-ray), text, time-series, and tabular data (EHR). Unlike previous narrowly focused reviews, our study encompasses a broad array of medical data modalities and explores various generative models. Our search strategy queries databases such as Scopus, PubMed, and ArXiv, focusing on recent works from January 2021 to November 2023, excluding reviews and perspectives. This period emphasizes recent advancements beyond GANs, which have been extensively covered previously. The survey reveals insights from three key aspects: (1) Synthesis applications and purpose of synthesis, (2) generation techniques, and (3) evaluation methods. It highlights clinically valid synthesis applications, demonstrating the potential of synthetic data to tackle diverse clinical requirements. While conditional models incorporating class labels, segmentation masks and image translations are prevalent, there is a gap in utilizing prior clinical knowledge and patient-specific context, suggesting a need for more personalized synthesis approaches and emphasizing the importance of tailoring generative approaches to the unique characteristics of medical data. Additionally, there is a significant gap in using synthetic data beyond augmentation, such as for validation and evaluation of downstream medical AI models. The survey uncovers that the lack of standardized evaluation methodologies tailored to medical images is a barrier to clinical application, underscoring the need for in-depth evaluation approaches, benchmarking, and comparative studies to promote openness and collaboration.
Histogram- and Diffusion-Based Medical Out-of-Distribution Detection
Oct 12, 2023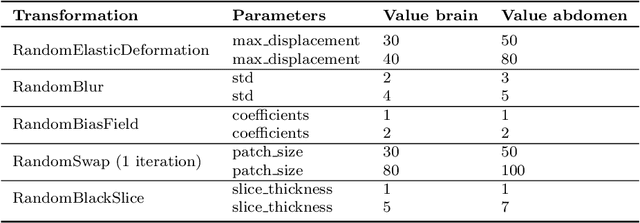
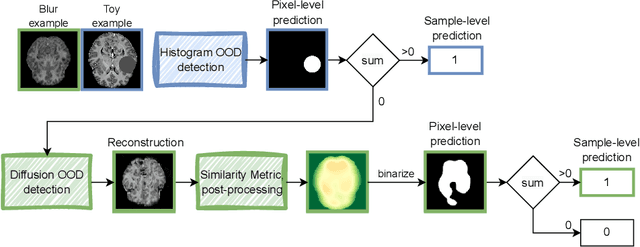
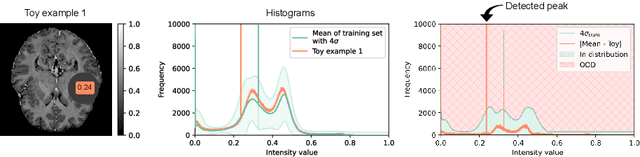

Abstract:Out-of-distribution (OOD) detection is crucial for the safety and reliability of artificial intelligence algorithms, especially in the medical domain. In the context of the Medical OOD (MOOD) detection challenge 2023, we propose a pipeline that combines a histogram-based method and a diffusion-based method. The histogram-based method is designed to accurately detect homogeneous anomalies in the toy examples of the challenge, such as blobs with constant intensity values. The diffusion-based method is based on one of the latest methods for unsupervised anomaly detection, called DDPM-OOD. We explore this method and propose extensive post-processing steps for pixel-level and sample-level anomaly detection on brain MRI and abdominal CT data provided by the challenge. Our results show that the proposed DDPM method is sensitive to blur and bias field samples, but faces challenges with anatomical deformation, black slice, and swapped patches. These findings suggest that further research is needed to improve the performance of DDPM for OOD detection in medical images.
A Deep Learning Approach Utilizing Covariance Matrix Analysis for the ISBI Edited MRS Reconstruction Challenge
Jun 05, 2023Abstract:This work proposes a method to accelerate the acquisition of high-quality edited magnetic resonance spectroscopy (MRS) scans using machine learning models taking the sample covariance matrix as input. The method is invariant to the number of transients and robust to noisy input data for both synthetic as well as in-vivo scenarios.
Pathology Synthesis of 3D Consistent Cardiac MR Im-ages Using 2D VAEs and GANs
Sep 09, 2022



Abstract:We propose a method for synthesizing cardiac MR images with plausible heart shapes and realistic appearances for the purpose of generating labeled data for deep-learning (DL) training. It breaks down the image synthesis into label deformation and label-to-image translation tasks. The former is achieved via latent space interpolation in a VAE model, while the latter is accomplished via a conditional GAN model. We devise an approach for label manipulation in the latent space of the trained VAE model, namely pathology synthesis, aiming to synthesize a series of pseudo-pathological synthetic subjects with characteristics of a desired heart disease. Furthermore, we propose to model the relationship between 2D slices in the latent space of the VAE via estimating the correlation coefficient matrix between the latent vectors and utilizing it to correlate elements of randomly drawn samples before decoding to image space. This simple yet effective approach results in generating 3D consistent subjects from 2D slice-by-slice generations. Such an approach could provide a solution to diversify and enrich the available database of cardiac MR images and to pave the way for the development of generalizable DL-based image analysis algorithms. The code will be available at https://github.com/sinaamirrajab/CardiacPathologySynthesis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge