Ruud J. G. van Sloun
Task-Based Adaptive Transmit Beamforming for Efficient Ultrasound Quantification
Jan 28, 2026Abstract:Wireless and wearable ultrasound devices promise to enable continuous ultrasound monitoring, but power consumption and data throughput remain critical challenges. Reducing the number of transmit events per second directly impacts both. We propose a task-based adaptive transmit beamforming method, formulated as a Bayesian active perception problem, that adaptively chooses where to scan in order to gain information about downstream quantitative measurements, avoiding redundant transmit events. Our proposed Task-Based Information Gain (TBIG) strategy applies to any differentiable downstream task function. When applied to recovering ventricular dimensions from echocardiograms, TBIG recovers accurate results using fewer than 2% of scan lines typically used, showing potential for large reductions in the power usage and data rates necessary for monitoring. Code is available at https://github.com/tue-bmd/task-based-ulsa.
Iterative Amortized Hierarchical VAE
Jan 22, 2026Abstract:In this paper we propose the Iterative Amortized Hierarchical Variational Autoencoder (IA-HVAE), which expands on amortized inference with a hybrid scheme containing an initial amortized guess and iterative refinement with decoder gradients. We achieve this by creating a linearly separable decoder in a transform domain (e.g. Fourier space), enabling real-time applications with very high model depths. The architectural change leads to a 35x speed-up for iterative inference with respect to the traditional HVAE. We show that our hybrid approach outperforms fully amortized and fully iterative equivalents in accuracy and speed respectively. Moreover, the IAHVAE shows improved reconstruction quality over a vanilla HVAE in inverse problems such as deblurring and denoising.
SVD-NO: Learning PDE Solution Operators with SVD Integral Kernels
Nov 13, 2025Abstract:Neural operators have emerged as a promising paradigm for learning solution operators of partial differential equa- tions (PDEs) directly from data. Existing methods, such as those based on Fourier or graph techniques, make strong as- sumptions about the structure of the kernel integral opera- tor, assumptions which may limit expressivity. We present SVD-NO, a neural operator that explicitly parameterizes the kernel by its singular-value decomposition (SVD) and then carries out the integral directly in the low-rank basis. Two lightweight networks learn the left and right singular func- tions, a diagonal parameter matrix learns the singular values, and a Gram-matrix regularizer enforces orthonormality. As SVD-NO approximates the full kernel, it obtains a high de- gree of expressivity. Furthermore, due to its low-rank struc- ture the computational complexity of applying the operator remains reasonable, leading to a practical system. In exten- sive evaluations on five diverse benchmark equations, SVD- NO achieves a new state of the art. In particular, SVD-NO provides greater performance gains on PDEs whose solutions are highly spatially variable. The code of this work is publicly available at https://github.com/2noamk/SVDNO.git.
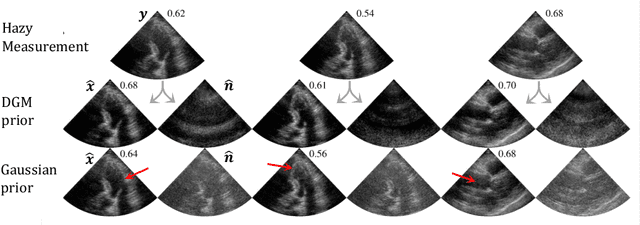
Semantic Diffusion Posterior Sampling for Cardiac Ultrasound Dehazing
Aug 24, 2025Abstract:Echocardiography plays a central role in cardiac imaging, offering dynamic views of the heart that are essential for diagnosis and monitoring. However, image quality can be significantly degraded by haze arising from multipath reverberations, particularly in difficult-to-image patients. In this work, we propose a semantic-guided, diffusion-based dehazing algorithm developed for the MICCAI Dehazing Echocardiography Challenge (DehazingEcho2025). Our method integrates a pixel-wise noise model, derived from semantic segmentation of hazy inputs into a diffusion posterior sampling framework guided by a generative prior trained on clean ultrasound data. Quantitative evaluation on the challenge dataset demonstrates strong performance across contrast and fidelity metrics. Code for the submitted algorithm is available at https://github.com/tristan-deep/semantic-diffusion-echo-dehazing.
Patient-Adaptive Focused Transmit Beamforming using Cognitive Ultrasound
Aug 12, 2025Abstract:Focused transmit beamforming is the most commonly used acquisition scheme for echocardiograms, but suffers from relatively low frame rates, and in 3D, even lower volume rates. Fast imaging based on unfocused transmits has disadvantages such as motion decorrelation and limited harmonic imaging capabilities. This work introduces a patient-adaptive focused transmit scheme that has the ability to drastically reduce the number of transmits needed to produce a high-quality ultrasound image. The method relies on posterior sampling with a temporal diffusion model to perceive and reconstruct the anatomy based on partial observations, while subsequently taking an action to acquire the most informative transmits. This active perception modality outperforms random and equispaced subsampling on the 2D EchoNet-Dynamic dataset and a 3D Philips dataset, where we actively select focused elevation planes. Furthermore, we show it achieves better performance in terms of generalized contrast-to-noise ratio when compared to the same number of diverging waves transmits on three in-house echocardiograms. Additionally, we can estimate ejection fraction using only 2% of the total transmits and show that the method is robust to outlier patients. Finally, our method can be run in real-time on GPU accelerators from 2023. The code is publicly available at https://tue-bmd.github.io/ulsa/
High Volume Rate 3D Ultrasound Reconstruction with Diffusion Models
May 28, 2025Abstract:Three-dimensional ultrasound enables real-time volumetric visualization of anatomical structures. Unlike traditional 2D ultrasound, 3D imaging reduces the reliance on precise probe orientation, potentially making ultrasound more accessible to clinicians with varying levels of experience and improving automated measurements and post-exam analysis. However, achieving both high volume rates and high image quality remains a significant challenge. While 3D diverging waves can provide high volume rates, they suffer from limited tissue harmonic generation and increased multipath effects, which degrade image quality. One compromise is to retain the focusing in elevation while leveraging unfocused diverging waves in the lateral direction to reduce the number of transmissions per elevation plane. Reaching the volume rates achieved by full 3D diverging waves, however, requires dramatically undersampling the number of elevation planes. Subsequently, to render the full volume, simple interpolation techniques are applied. This paper introduces a novel approach to 3D ultrasound reconstruction from a reduced set of elevation planes by employing diffusion models (DMs) to achieve increased spatial and temporal resolution. We compare both traditional and supervised deep learning-based interpolation methods on a 3D cardiac ultrasound dataset. Our results show that DM-based reconstruction consistently outperforms the baselines in image quality and downstream task performance. Additionally, we accelerate inference by leveraging the temporal consistency inherent to ultrasound sequences. Finally, we explore the robustness of the proposed method by exploiting the probabilistic nature of diffusion posterior sampling to quantify reconstruction uncertainty and demonstrate improved recall on out-of-distribution data with synthetic anomalies under strong subsampling.
EOTNet: Deep Memory Aided Bayesian Filter for Extended Object Tracking
May 24, 2025Abstract:Extended object tracking methods based on random matrices, founded on Bayesian filters, have been able to achieve efficient recursive processes while jointly estimating the kinematic states and extension of the targets. Existing random matrix approaches typically assume that the evolution of state and extension follows a first-order Markov process, where the current estimate of the target depends solely on the previous moment. However, in real-world scenarios, this assumption fails because the evolution of states and extension is usually non-Markovian. In this paper, we introduce a novel extended object tracking method: a Bayesian recursive neural network assisted by deep memory. Initially, we propose an equivalent model under a non-Markovian assumption and derive the implementation of its Bayesian filtering framework. Thereafter, Gaussian approximation and moment matching are employed to derive the analytical solution for the proposed Bayesian filtering framework. Finally, based on the closed-form solution, we design an end-to-end trainable Bayesian recursive neural network for extended object tracking. Experiment results on simulated and real-world datasets show that the proposed methods outperforms traditional extended object tracking methods and state-of-the-art deep learning approaches.
Physics-Informed Sylvester Normalizing Flows for Bayesian Inference in Magnetic Resonance Spectroscopy
May 06, 2025Abstract:Magnetic resonance spectroscopy (MRS) is a non-invasive technique to measure the metabolic composition of tissues, offering valuable insights into neurological disorders, tumor detection, and other metabolic dysfunctions. However, accurate metabolite quantification is hindered by challenges such as spectral overlap, low signal-to-noise ratio, and various artifacts. Traditional methods like linear-combination modeling are susceptible to ambiguities and commonly only provide a theoretical lower bound on estimation accuracy in the form of the Cram\'er-Rao bound. This work introduces a Bayesian inference framework using Sylvester normalizing flows (SNFs) to approximate posterior distributions over metabolite concentrations, enhancing quantification reliability. A physics-based decoder incorporates prior knowledge of MRS signal formation, ensuring realistic distribution representations. We validate the method on simulated 7T proton MRS data, demonstrating accurate metabolite quantification, well-calibrated uncertainties, and insights into parameter correlations and multi-modal distributions.
Deep Generative Models for Bayesian Inference on High-Rate Sensor Data: Applications in Automotive Radar and Medical Imaging
Apr 16, 2025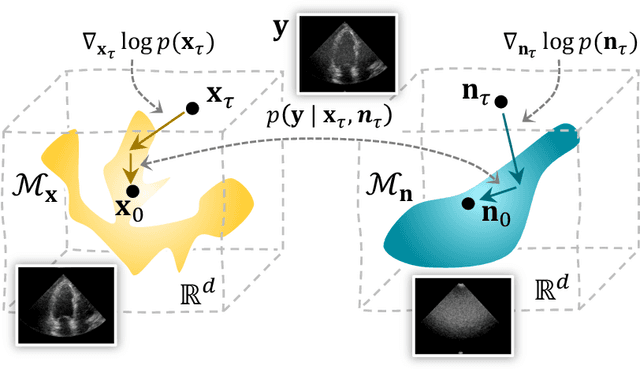
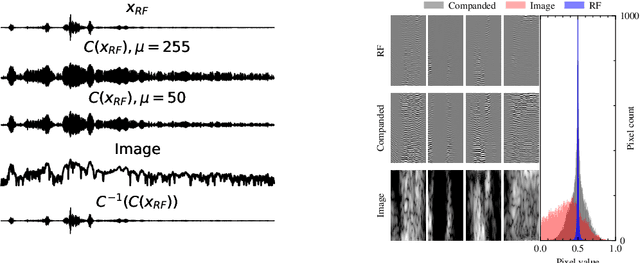
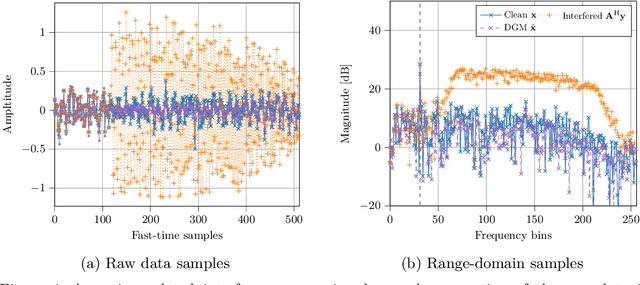
Abstract:Deep generative models have been studied and developed primarily in the context of natural images and computer vision. This has spurred the development of (Bayesian) methods that use these generative models for inverse problems in image restoration, such as denoising, inpainting, and super-resolution. In recent years, generative modeling for Bayesian inference on sensory data has also gained traction. Nevertheless, the direct application of generative modeling techniques initially designed for natural images on raw sensory data is not straightforward, requiring solutions that deal with high dynamic range signals acquired from multiple sensors or arrays of sensors that interfere with each other, and that typically acquire data at a very high rate. Moreover, the exact physical data-generating process is often complex or unknown. As a consequence, approximate models are used, resulting in discrepancies between model predictions and the observations that are non-Gaussian, in turn complicating the Bayesian inverse problem. Finally, sensor data is often used in real-time processing or decision-making systems, imposing stringent requirements on, e.g., latency and throughput. In this paper, we will discuss some of these challenges and offer approaches to address them, all in the context of high-rate real-time sensing applications in automotive radar and medical imaging.
Deep Variational Sequential Monte Carlo for High-Dimensional Observations
Jan 10, 2025Abstract:Sequential Monte Carlo (SMC), or particle filtering, is widely used in nonlinear state-space systems, but its performance often suffers from poorly approximated proposal and state-transition distributions. This work introduces a differentiable particle filter that leverages the unsupervised variational SMC objective to parameterize the proposal and transition distributions with a neural network, designed to learn from high-dimensional observations. Experimental results demonstrate that our approach outperforms established baselines in tracking the challenging Lorenz attractor from high-dimensional and partial observations. Furthermore, an evidence lower bound based evaluation indicates that our method offers a more accurate representation of the posterior distribution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge