Marcel Breeuwer
SAM-Fed: SAM-Guided Federated Semi-Supervised Learning for Medical Image Segmentation
Nov 18, 2025Abstract:Medical image segmentation is clinically important, yet data privacy and the cost of expert annotation limit the availability of labeled data. Federated semi-supervised learning (FSSL) offers a solution but faces two challenges: pseudo-label reliability depends on the strength of local models, and client devices often require compact or heterogeneous architectures due to limited computational resources. These constraints reduce the quality and stability of pseudo-labels, while large models, though more accurate, cannot be trained or used for routine inference on client devices. We propose SAM-Fed, a federated semi-supervised framework that leverages a high-capacity segmentation foundation model to guide lightweight clients during training. SAM-Fed combines dual knowledge distillation with an adaptive agreement mechanism to refine pixel-level supervision. Experiments on skin lesion and polyp segmentation across homogeneous and heterogeneous settings show that SAM-Fed consistently outperforms state-of-the-art FSSL methods.
Scaling up self-supervised learning for improved surgical foundation models
Jan 16, 2025Abstract:Foundation models have revolutionized computer vision by achieving vastly superior performance across diverse tasks through large-scale pretraining on extensive datasets. However, their application in surgical computer vision has been limited. This study addresses this gap by introducing SurgeNetXL, a novel surgical foundation model that sets a new benchmark in surgical computer vision. Trained on the largest reported surgical dataset to date, comprising over 4.7 million video frames, SurgeNetXL achieves consistent top-tier performance across six datasets spanning four surgical procedures and three tasks, including semantic segmentation, phase recognition, and critical view of safety (CVS) classification. Compared with the best-performing surgical foundation models, SurgeNetXL shows mean improvements of 2.4, 9.0, and 12.6 percent for semantic segmentation, phase recognition, and CVS classification, respectively. Additionally, SurgeNetXL outperforms the best-performing ImageNet-based variants by 14.4, 4.0, and 1.6 percent in the respective tasks. In addition to advancing model performance, this study provides key insights into scaling pretraining datasets, extending training durations, and optimizing model architectures specifically for surgical computer vision. These findings pave the way for improved generalizability and robustness in data-scarce scenarios, offering a comprehensive framework for future research in this domain. All models and a subset of the SurgeNetXL dataset, including over 2 million video frames, are publicly available at: https://github.com/TimJaspers0801/SurgeNet.
Benchmarking and Enhancing Surgical Phase Recognition Models for Robotic-Assisted Esophagectomy
Dec 05, 2024



Abstract:Robotic-assisted minimally invasive esophagectomy (RAMIE) is a recognized treatment for esophageal cancer, offering better patient outcomes compared to open surgery and traditional minimally invasive surgery. RAMIE is highly complex, spanning multiple anatomical areas and involving repetitive phases and non-sequential phase transitions. Our goal is to leverage deep learning for surgical phase recognition in RAMIE to provide intraoperative support to surgeons. To achieve this, we have developed a new surgical phase recognition dataset comprising 27 videos. Using this dataset, we conducted a comparative analysis of state-of-the-art surgical phase recognition models. To more effectively capture the temporal dynamics of this complex procedure, we developed a novel deep learning model featuring an encoder-decoder structure with causal hierarchical attention, which demonstrates superior performance compared to existing models.
Benchmarking Pretrained Attention-based Models for Real-Time Recognition in Robot-Assisted Esophagectomy
Dec 04, 2024Abstract:Esophageal cancer is among the most common types of cancer worldwide. It is traditionally treated using open esophagectomy, but in recent years, robot-assisted minimally invasive esophagectomy (RAMIE) has emerged as a promising alternative. However, robot-assisted surgery can be challenging for novice surgeons, as they often suffer from a loss of spatial orientation. Computer-aided anatomy recognition holds promise for improving surgical navigation, but research in this area remains limited. In this study, we developed a comprehensive dataset for semantic segmentation in RAMIE, featuring the largest collection of vital anatomical structures and surgical instruments to date. Handling this diverse set of classes presents challenges, including class imbalance and the recognition of complex structures such as nerves. This study aims to understand the challenges and limitations of current state-of-the-art algorithms on this novel dataset and problem. Therefore, we benchmarked eight real-time deep learning models using two pretraining datasets. We assessed both traditional and attention-based networks, hypothesizing that attention-based networks better capture global patterns and address challenges such as occlusion caused by blood or other tissues. The benchmark includes our RAMIE dataset and the publicly available CholecSeg8k dataset, enabling a thorough assessment of surgical segmentation tasks. Our findings indicate that pretraining on ADE20k, a dataset for semantic segmentation, is more effective than pretraining on ImageNet. Furthermore, attention-based models outperform traditional convolutional neural networks, with SegNeXt and Mask2Former achieving higher Dice scores, and Mask2Former additionally excelling in average symmetric surface distance.
Generative AI for Synthetic Data Across Multiple Medical Modalities: A Systematic Review of Recent Developments and Challenges
Jul 02, 2024


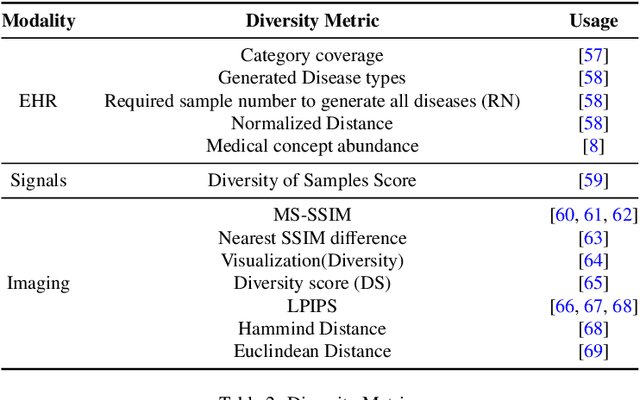
Abstract:This paper presents a comprehensive systematic review of generative models (GANs, VAEs, DMs, and LLMs) used to synthesize various medical data types, including imaging (dermoscopic, mammographic, ultrasound, CT, MRI, and X-ray), text, time-series, and tabular data (EHR). Unlike previous narrowly focused reviews, our study encompasses a broad array of medical data modalities and explores various generative models. Our search strategy queries databases such as Scopus, PubMed, and ArXiv, focusing on recent works from January 2021 to November 2023, excluding reviews and perspectives. This period emphasizes recent advancements beyond GANs, which have been extensively covered previously. The survey reveals insights from three key aspects: (1) Synthesis applications and purpose of synthesis, (2) generation techniques, and (3) evaluation methods. It highlights clinically valid synthesis applications, demonstrating the potential of synthetic data to tackle diverse clinical requirements. While conditional models incorporating class labels, segmentation masks and image translations are prevalent, there is a gap in utilizing prior clinical knowledge and patient-specific context, suggesting a need for more personalized synthesis approaches and emphasizing the importance of tailoring generative approaches to the unique characteristics of medical data. Additionally, there is a significant gap in using synthetic data beyond augmentation, such as for validation and evaluation of downstream medical AI models. The survey uncovers that the lack of standardized evaluation methodologies tailored to medical images is a barrier to clinical application, underscoring the need for in-depth evaluation approaches, benchmarking, and comparative studies to promote openness and collaboration.
A Deep Learning Approach Utilizing Covariance Matrix Analysis for the ISBI Edited MRS Reconstruction Challenge
Jun 05, 2023Abstract:This work proposes a method to accelerate the acquisition of high-quality edited magnetic resonance spectroscopy (MRS) scans using machine learning models taking the sample covariance matrix as input. The method is invariant to the number of transients and robust to noisy input data for both synthetic as well as in-vivo scenarios.
Pathology Synthesis of 3D Consistent Cardiac MR Im-ages Using 2D VAEs and GANs
Sep 09, 2022



Abstract:We propose a method for synthesizing cardiac MR images with plausible heart shapes and realistic appearances for the purpose of generating labeled data for deep-learning (DL) training. It breaks down the image synthesis into label deformation and label-to-image translation tasks. The former is achieved via latent space interpolation in a VAE model, while the latter is accomplished via a conditional GAN model. We devise an approach for label manipulation in the latent space of the trained VAE model, namely pathology synthesis, aiming to synthesize a series of pseudo-pathological synthetic subjects with characteristics of a desired heart disease. Furthermore, we propose to model the relationship between 2D slices in the latent space of the VAE via estimating the correlation coefficient matrix between the latent vectors and utilizing it to correlate elements of randomly drawn samples before decoding to image space. This simple yet effective approach results in generating 3D consistent subjects from 2D slice-by-slice generations. Such an approach could provide a solution to diversify and enrich the available database of cardiac MR images and to pave the way for the development of generalizable DL-based image analysis algorithms. The code will be available at https://github.com/sinaamirrajab/CardiacPathologySynthesis.
sim2real: Cardiac MR Image Simulation-to-Real Translation via Unsupervised GANs
Aug 09, 2022



Abstract:There has been considerable interest in the MR physics-based simulation of a database of virtual cardiac MR images for the development of deep-learning analysis networks. However, the employment of such a database is limited or shows suboptimal performance due to the realism gap, missing textures, and the simplified appearance of simulated images. In this work we 1) provide image simulation on virtual XCAT subjects with varying anatomies, and 2) propose sim2real translation network to improve image realism. Our usability experiments suggest that sim2real data exhibits a good potential to augment training data and boost the performance of a segmentation algorithm.
Optimized Automated Cardiac MR Scar Quantification with GAN-Based Data Augmentation
Sep 27, 2021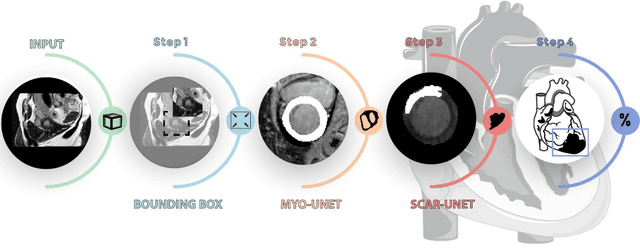
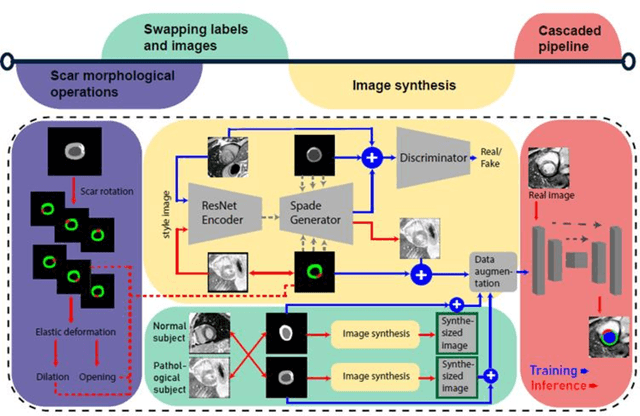
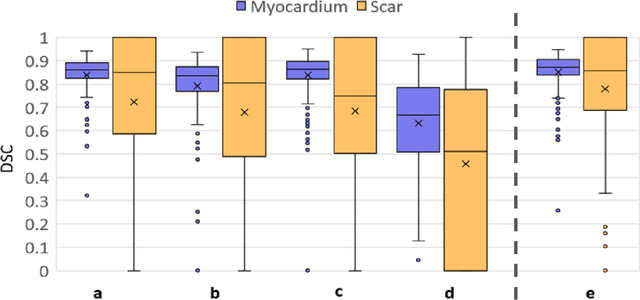
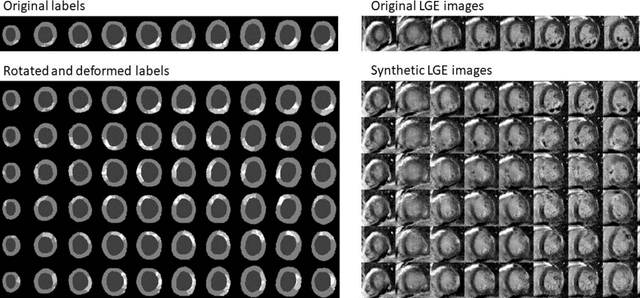
Abstract:Background: The clinical utility of late gadolinium enhancement (LGE) cardiac MRI is limited by the lack of standardization, and time-consuming postprocessing. In this work, we tested the hypothesis that a cascaded deep learning pipeline trained with augmentation by synthetically generated data would improve model accuracy and robustness for automated scar quantification. Methods: A cascaded pipeline consisting of three consecutive neural networks is proposed, starting with a bounding box regression network to identify a region of interest around the left ventricular (LV) myocardium. Two further nnU-Net models are then used to segment the myocardium and, if present, scar. The models were trained on the data from the EMIDEC challenge, supplemented with an extensive synthetic dataset generated with a conditional GAN. Results: The cascaded pipeline significantly outperformed a single nnU-Net directly segmenting both the myocardium (mean Dice similarity coefficient (DSC) (standard deviation (SD)): 0.84 (0.09) vs 0.63 (0.20), p < 0.01) and scar (DSC: 0.72 (0.34) vs 0.46 (0.39), p < 0.01) on a per-slice level. The inclusion of the synthetic data as data augmentation during training improved the scar segmentation DSC by 0.06 (p < 0.01). The mean DSC per-subject on the challenge test set, for the cascaded pipeline augmented by synthetic generated data, was 0.86 (0.03) and 0.67 (0.29) for myocardium and scar, respectively. Conclusion: A cascaded deep learning-based pipeline trained with augmentation by synthetically generated data leads to myocardium and scar segmentations that are similar to the manual operator, and outperforms direct segmentation without the synthetic images.
Physics-informed neural networks for myocardial perfusion MRI quantification
Dec 07, 2020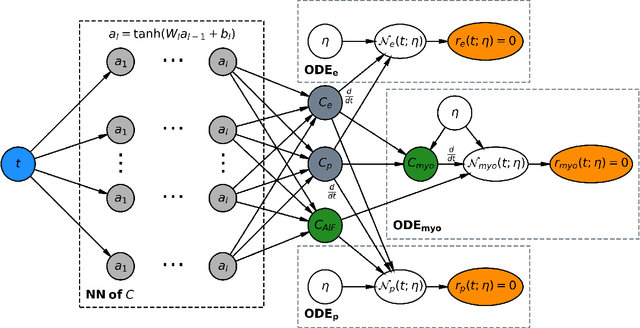
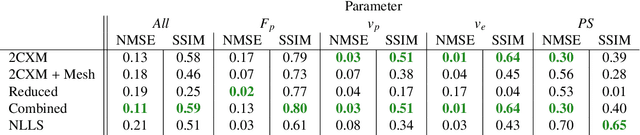
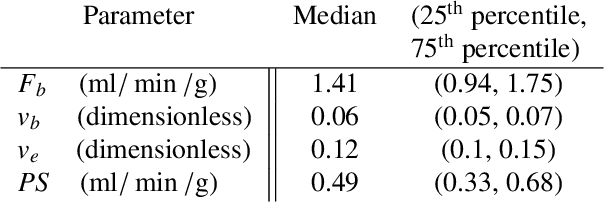
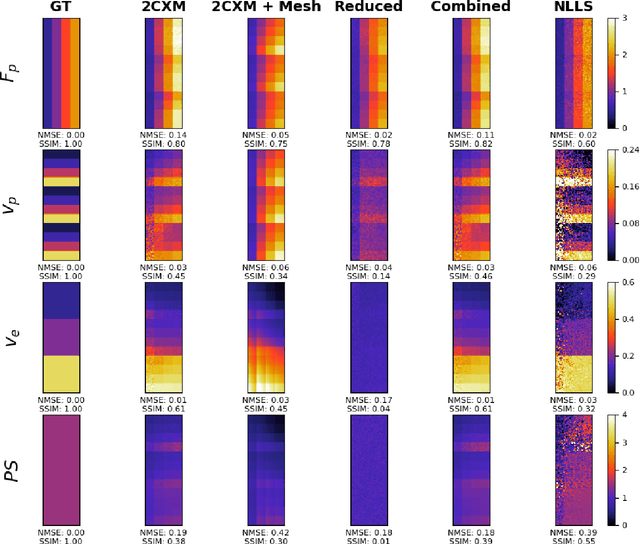
Abstract:Tracer-kinetic models allow for the quantification of kinetic parameters such as blood flow from dynamic contrast-enhanced magnetic resonance (MR) images. Fitting the observed data with multi-compartment exchange models is desirable, as they are physiologically plausible and resolve directly for blood flow and microvascular function. However, the reliability of model fitting is limited by the low signal-to-noise ratio, temporal resolution, and acquisition length. This may result in inaccurate parameter estimates. This study introduces physics-informed neural networks (PINNs) as a means to perform myocardial perfusion MR quantification, which provides a versatile scheme for the inference of kinetic parameters. These neural networks can be trained to fit the observed perfusion MR data while respecting the underlying physical conservation laws described by a multi-compartment exchange model. Here, we provide a framework for the implementation of PINNs in myocardial perfusion MR. The approach is validated both in silico and in vivo. In the in silico study, an overall reduction in mean-squared error with the ground-truth parameters was observed compared to a standard non-linear least squares fitting approach. The in vivo study demonstrates that the method produces parameter values comparable to those previously found in literature, as well as providing parameter maps which match the clinical diagnosis of patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge