Rudolf L. M. van Herten
Attenuation artifact detection and severity classification in intracoronary OCT using mixed image representations
Mar 07, 2025Abstract:In intracoronary optical coherence tomography (OCT), blood residues and gas bubbles cause attenuation artifacts that can obscure critical vessel structures. The presence and severity of these artifacts may warrant re-acquisition, prolonging procedure time and increasing use of contrast agent. Accurate detection of these artifacts can guide targeted re-acquisition, reducing the amount of repeated scans needed to achieve diagnostically viable images. However, the highly heterogeneous appearance of these artifacts poses a challenge for the automated detection of the affected image regions. To enable automatic detection of the attenuation artifacts caused by blood residues and gas bubbles based on their severity, we propose a convolutional neural network that performs classification of the attenuation lines (A-lines) into three classes: no artifact, mild artifact and severe artifact. Our model extracts and merges features from OCT images in both Cartesian and polar coordinates, where each column of the image represents an A-line. Our method detects the presence of attenuation artifacts in OCT frames reaching F-scores of 0.77 and 0.94 for mild and severe artifacts, respectively. The inference time over a full OCT scan is approximately 6 seconds. Our experiments show that analysis of images represented in both Cartesian and polar coordinate systems outperforms the analysis in polar coordinates only, suggesting that these representations contain complementary features. This work lays the foundation for automated artifact assessment and image acquisition guidance in intracoronary OCT imaging.
World of Forms: Deformable Geometric Templates for One-Shot Surface Meshing in Coronary CT Angiography
Sep 18, 2024Abstract:Deep learning-based medical image segmentation and surface mesh generation typically involve a sequential pipeline from image to segmentation to meshes, often requiring large training datasets while making limited use of prior geometric knowledge. This may lead to topological inconsistencies and suboptimal performance in low-data regimes. To address these challenges, we propose a data-efficient deep learning method for direct 3D anatomical object surface meshing using geometric priors. Our approach employs a multi-resolution graph neural network that operates on a prior geometric template which is deformed to fit object boundaries of interest. We show how different templates may be used for the different surface meshing targets, and introduce a novel masked autoencoder pretraining strategy for 3D spherical data. The proposed method outperforms nnUNet in a one-shot setting for segmentation of the pericardium, left ventricle (LV) cavity and the LV myocardium. Similarly, the method outperforms other lumen segmentation operating on multi-planar reformatted images. Results further indicate that mesh quality is on par with or improves upon marching cubes post-processing of voxel mask predictions, while remaining flexible in the choice of mesh triangulation prior, thus paving the way for more accurate and topologically consistent 3D medical object surface meshing.
Automatic Coronary Artery Plaque Quantification and CAD-RADS Prediction using Mesh Priors
Oct 17, 2023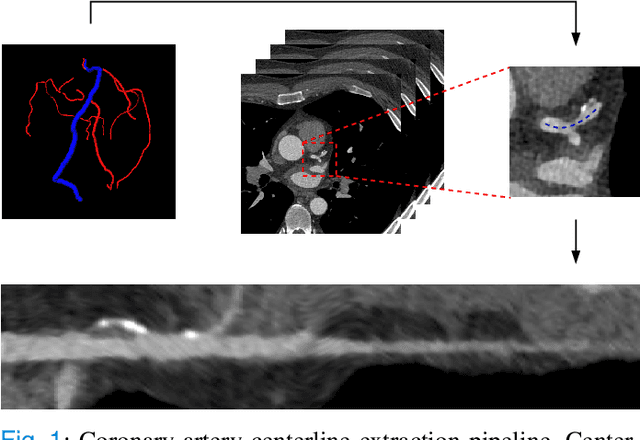
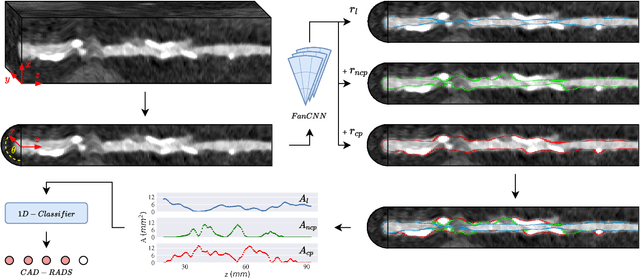
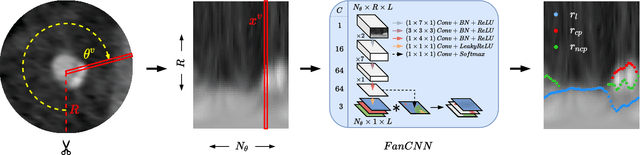
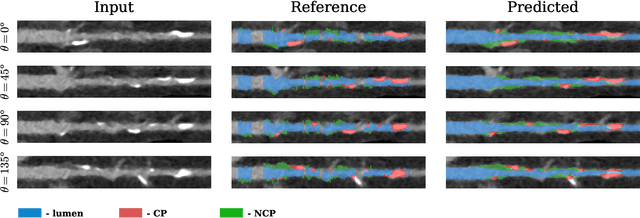
Abstract:Coronary artery disease (CAD) remains the leading cause of death worldwide. Patients with suspected CAD undergo coronary CT angiography (CCTA) to evaluate the risk of cardiovascular events and determine the treatment. Clinical analysis of coronary arteries in CCTA comprises the identification of atherosclerotic plaque, as well as the grading of any coronary artery stenosis typically obtained through the CAD-Reporting and Data System (CAD-RADS). This requires analysis of the coronary lumen and plaque. While voxel-wise segmentation is a commonly used approach in various segmentation tasks, it does not guarantee topologically plausible shapes. To address this, in this work, we propose to directly infer surface meshes for coronary artery lumen and plaque based on a centerline prior and use it in the downstream task of CAD-RADS scoring. The method is developed and evaluated using a total of 2407 CCTA scans. Our method achieved lesion-wise volume intraclass correlation coefficients of 0.98, 0.79, and 0.85 for calcified, non-calcified, and total plaque volume respectively. Patient-level CAD-RADS categorization was evaluated on a representative hold-out test set of 300 scans, for which the achieved linearly weighted kappa ($\kappa$) was 0.75. CAD-RADS categorization on the set of 658 scans from another hospital and scanner led to a $\kappa$ of 0.71. The results demonstrate that direct inference of coronary artery meshes for lumen and plaque is feasible, and allows for the automated prediction of routinely performed CAD-RADS categorization.
Physics-informed neural networks for myocardial perfusion MRI quantification
Dec 07, 2020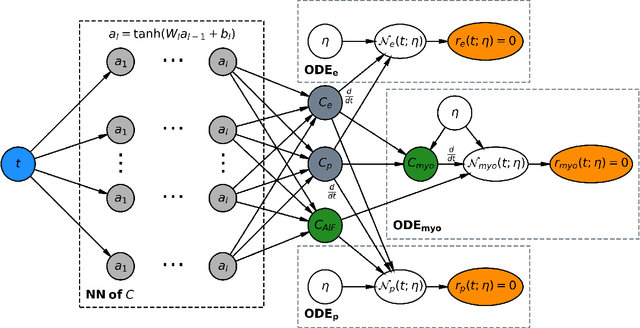
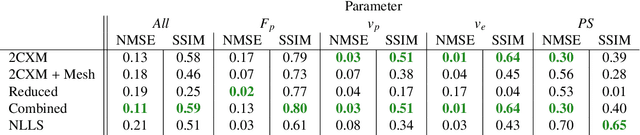
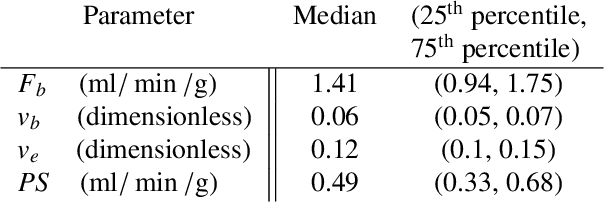
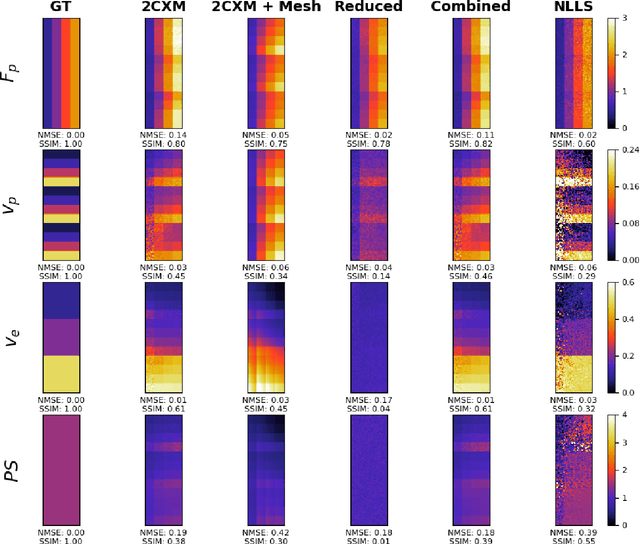
Abstract:Tracer-kinetic models allow for the quantification of kinetic parameters such as blood flow from dynamic contrast-enhanced magnetic resonance (MR) images. Fitting the observed data with multi-compartment exchange models is desirable, as they are physiologically plausible and resolve directly for blood flow and microvascular function. However, the reliability of model fitting is limited by the low signal-to-noise ratio, temporal resolution, and acquisition length. This may result in inaccurate parameter estimates. This study introduces physics-informed neural networks (PINNs) as a means to perform myocardial perfusion MR quantification, which provides a versatile scheme for the inference of kinetic parameters. These neural networks can be trained to fit the observed perfusion MR data while respecting the underlying physical conservation laws described by a multi-compartment exchange model. Here, we provide a framework for the implementation of PINNs in myocardial perfusion MR. The approach is validated both in silico and in vivo. In the in silico study, an overall reduction in mean-squared error with the ground-truth parameters was observed compared to a standard non-linear least squares fitting approach. The in vivo study demonstrates that the method produces parameter values comparable to those previously found in literature, as well as providing parameter maps which match the clinical diagnosis of patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge