Jelmer M. Wolterink
Consistent View Alignment Improves Foundation Models for 3D Medical Image Segmentation
Sep 17, 2025Abstract:Many recent approaches in representation learning implicitly assume that uncorrelated views of a data point are sufficient to learn meaningful representations for various downstream tasks. In this work, we challenge this assumption and demonstrate that meaningful structure in the latent space does not emerge naturally. Instead, it must be explicitly induced. We propose a method that aligns representations from different views of the data to align complementary information without inducing false positives. Our experiments show that our proposed self-supervised learning method, Consistent View Alignment, improves performance for downstream tasks, highlighting the critical role of structured view alignment in learning effective representations. Our method achieved first and second place in the MICCAI 2025 SSL3D challenge when using a Primus vision transformer and ResEnc convolutional neural network, respectively. The code and pretrained model weights are released at https://github.com/Tenbatsu24/LatentCampus.
GReAT: leveraging geometric artery data to improve wall shear stress assessment
Aug 26, 2025Abstract:Leveraging big data for patient care is promising in many medical fields such as cardiovascular health. For example, hemodynamic biomarkers like wall shear stress could be assessed from patient-specific medical images via machine learning algorithms, bypassing the need for time-intensive computational fluid simulation. However, it is extremely challenging to amass large-enough datasets to effectively train such models. We could address this data scarcity by means of self-supervised pre-training and foundations models given large datasets of geometric artery models. In the context of coronary arteries, leveraging learned representations to improve hemodynamic biomarker assessment has not yet been well studied. In this work, we address this gap by investigating whether a large dataset (8449 shapes) consisting of geometric models of 3D blood vessels can benefit wall shear stress assessment in coronary artery models from a small-scale clinical trial (49 patients). We create a self-supervised target for the 3D blood vessels by computing the heat kernel signature, a quantity obtained via Laplacian eigenvectors, which captures the very essence of the shapes. We show how geometric representations learned from this datasets can boost segmentation of coronary arteries into regions of low, mid and high (time-averaged) wall shear stress even when trained on limited data.
Beyond Pixels: Medical Image Quality Assessment with Implicit Neural Representations
Aug 07, 2025Abstract:Artifacts pose a significant challenge in medical imaging, impacting diagnostic accuracy and downstream analysis. While image-based approaches for detecting artifacts can be effective, they often rely on preprocessing methods that can lead to information loss and high-memory-demand medical images, thereby limiting the scalability of classification models. In this work, we propose the use of implicit neural representations (INRs) for image quality assessment. INRs provide a compact and continuous representation of medical images, naturally handling variations in resolution and image size while reducing memory overhead. We develop deep weight space networks, graph neural networks, and relational attention transformers that operate on INRs to achieve image quality assessment. Our method is evaluated on the ACDC dataset with synthetically generated artifact patterns, demonstrating its effectiveness in assessing image quality while achieving similar performance with fewer parameters.
Wall Shear Stress Estimation in Abdominal Aortic Aneurysms: Towards Generalisable Neural Surrogate Models
Jul 30, 2025Abstract:Abdominal aortic aneurysms (AAAs) are pathologic dilatations of the abdominal aorta posing a high fatality risk upon rupture. Studying AAA progression and rupture risk often involves in-silico blood flow modelling with computational fluid dynamics (CFD) and extraction of hemodynamic factors like time-averaged wall shear stress (TAWSS) or oscillatory shear index (OSI). However, CFD simulations are known to be computationally demanding. Hence, in recent years, geometric deep learning methods, operating directly on 3D shapes, have been proposed as compelling surrogates, estimating hemodynamic parameters in just a few seconds. In this work, we propose a geometric deep learning approach to estimating hemodynamics in AAA patients, and study its generalisability to common factors of real-world variation. We propose an E(3)-equivariant deep learning model utilising novel robust geometrical descriptors and projective geometric algebra. Our model is trained to estimate transient WSS using a dataset of CT scans of 100 AAA patients, from which lumen geometries are extracted and reference CFD simulations with varying boundary conditions are obtained. Results show that the model generalizes well within the distribution, as well as to the external test set. Moreover, the model can accurately estimate hemodynamics across geometry remodelling and changes in boundary conditions. Furthermore, we find that a trained model can be applied to different artery tree topologies, where new and unseen branches are added during inference. Finally, we find that the model is to a large extent agnostic to mesh resolution. These results show the accuracy and generalisation of the proposed model, and highlight its potential to contribute to hemodynamic parameter estimation in clinical practice.
Geometric deep learning for local growth prediction on abdominal aortic aneurysm surfaces
Jun 11, 2025Abstract:Abdominal aortic aneurysms (AAAs) are progressive focal dilatations of the abdominal aorta. AAAs may rupture, with a survival rate of only 20\%. Current clinical guidelines recommend elective surgical repair when the maximum AAA diameter exceeds 55 mm in men or 50 mm in women. Patients that do not meet these criteria are periodically monitored, with surveillance intervals based on the maximum AAA diameter. However, this diameter does not take into account the complex relation between the 3D AAA shape and its growth, making standardized intervals potentially unfit. Personalized AAA growth predictions could improve monitoring strategies. We propose to use an SE(3)-symmetric transformer model to predict AAA growth directly on the vascular model surface enriched with local, multi-physical features. In contrast to other works which have parameterized the AAA shape, this representation preserves the vascular surface's anatomical structure and geometric fidelity. We train our model using a longitudinal dataset of 113 computed tomography angiography (CTA) scans of 24 AAA patients at irregularly sampled intervals. After training, our model predicts AAA growth to the next scan moment with a median diameter error of 1.18 mm. We further demonstrate our model's utility to identify whether a patient will become eligible for elective repair within two years (acc = 0.93). Finally, we evaluate our model's generalization on an external validation set consisting of 25 CTAs from 7 AAA patients from a different hospital. Our results show that local directional AAA growth prediction from the vascular surface is feasible and may contribute to personalized surveillance strategies.
Data-Agnostic Augmentations for Unknown Variations: Out-of-Distribution Generalisation in MRI Segmentation
May 15, 2025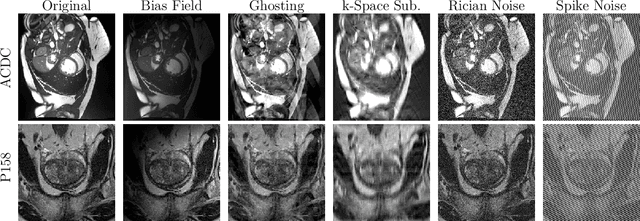
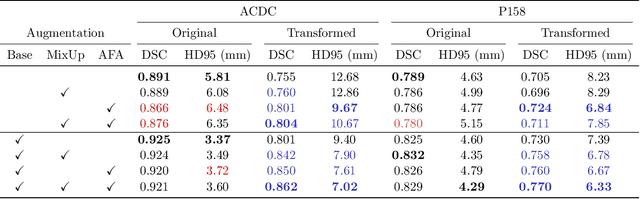
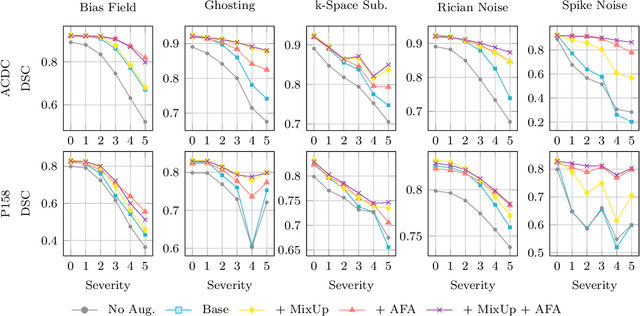
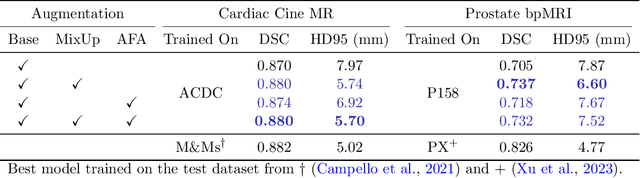
Abstract:Medical image segmentation models are often trained on curated datasets, leading to performance degradation when deployed in real-world clinical settings due to mismatches between training and test distributions. While data augmentation techniques are widely used to address these challenges, traditional visually consistent augmentation strategies lack the robustness needed for diverse real-world scenarios. In this work, we systematically evaluate alternative augmentation strategies, focusing on MixUp and Auxiliary Fourier Augmentation. These methods mitigate the effects of multiple variations without explicitly targeting specific sources of distribution shifts. We demonstrate how these techniques significantly improve out-of-distribution generalization and robustness to imaging variations across a wide range of transformations in cardiac cine MRI and prostate MRI segmentation. We quantitatively find that these augmentation methods enhance learned feature representations by promoting separability and compactness. Additionally, we highlight how their integration into nnU-Net training pipelines provides an easy-to-implement, effective solution for enhancing the reliability of medical segmentation models in real-world applications.
Steerable Anatomical Shape Synthesis with Implicit Neural Representations
Apr 04, 2025
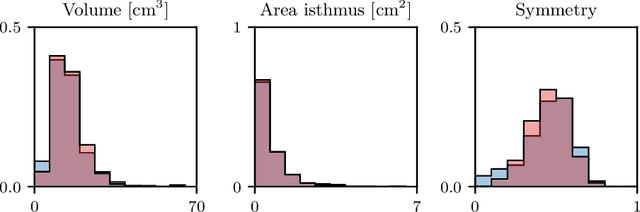
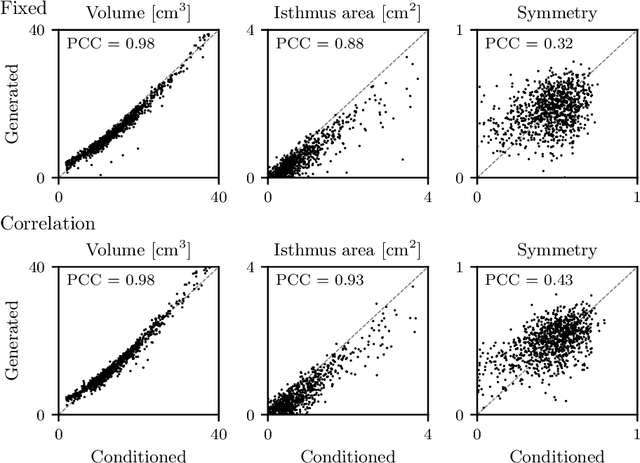
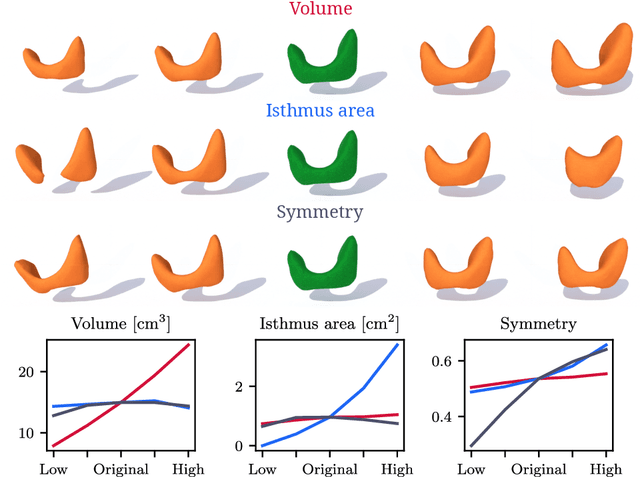
Abstract:Generative modeling of anatomical structures plays a crucial role in virtual imaging trials, which allow researchers to perform studies without the costs and constraints inherent to in vivo and phantom studies. For clinical relevance, generative models should allow targeted control to simulate specific patient populations rather than relying on purely random sampling. In this work, we propose a steerable generative model based on implicit neural representations. Implicit neural representations naturally support topology changes, making them well-suited for anatomical structures with varying topology, such as the thyroid. Our model learns a disentangled latent representation, enabling fine-grained control over shape variations. Evaluation includes reconstruction accuracy and anatomical plausibility. Our results demonstrate that the proposed model achieves high-quality shape generation while enabling targeted anatomical modifications.
Active Learning for Deep Learning-Based Hemodynamic Parameter Estimation
Mar 05, 2025Abstract:Hemodynamic parameters such as pressure and wall shear stress play an important role in diagnosis, prognosis, and treatment planning in cardiovascular diseases. These parameters can be accurately computed using computational fluid dynamics (CFD), but CFD is computationally intensive. Hence, deep learning methods have been adopted as a surrogate to rapidly estimate CFD outcomes. A drawback of such data-driven models is the need for time-consuming reference CFD simulations for training. In this work, we introduce an active learning framework to reduce the number of CFD simulations required for the training of surrogate models, lowering the barriers to their deployment in new applications. We propose three distinct querying strategies to determine for which unlabeled samples CFD simulations should be obtained. These querying strategies are based on geometrical variance, ensemble uncertainty, and adherence to the physics governing fluid dynamics. We benchmark these methods on velocity field estimation in synthetic coronary artery bifurcations and find that they allow for substantial reductions in annotation cost. Notably, we find that our strategies reduce the number of samples required by up to 50% and make the trained models more robust to difficult cases. Our results show that active learning is a feasible strategy to increase the potential of deep learning-based CFD surrogates.
Waveform-Specific Performance of Deep Learning-Based Super-Resolution for Ultrasound Contrast Imaging
Jan 30, 2025



Abstract:Resolving arterial flows is essential for understanding cardiovascular pathologies, improving diagnosis, and monitoring patient condition. Ultrasound contrast imaging uses microbubbles to enhance the scattering of the blood pool, allowing for real-time visualization of blood flow. Recent developments in vector flow imaging further expand the imaging capabilities of ultrasound by temporally resolving fast arterial flow. The next obstacle to overcome is the lack of spatial resolution. Super-resolved ultrasound images can be obtained by deconvolving radiofrequency (RF) signals before beamforming, breaking the link between resolution and pulse duration. Convolutional neural networks (CNNs) can be trained to locally estimate the deconvolution kernel and consequently super-localize the microbubbles directly within the RF signal. However, microbubble contrast is highly nonlinear, and the potential of CNNs in microbubble localization has not yet been fully exploited. Assessing deep learning-based deconvolution performance for non-trivial imaging pulses is therefore essential for successful translation to a practical setting, where the signal-to-noise ratio is limited, and transmission schemes should comply with safety guidelines. In this study, we train CNNs to deconvolve RF signals and localize the microbubbles driven by harmonic pulses, chirps, or delay-encoded pulse trains. Furthermore, we discuss potential hurdles for in-vitro and in-vivo super-resolution by presenting preliminary experimental results. We find that, whereas the CNNs can accurately localize microbubbles for all pulses, a short imaging pulse offers the best performance in noise-free conditions. However, chirps offer a comparable performance without noise, but are more robust to noise and outperform all other pulses in low-signal-to-noise ratio conditions.
Deep vectorised operators for pulsatile hemodynamics estimation in coronary arteries from a steady-state prior
Oct 15, 2024



Abstract:Cardiovascular hemodynamic fields provide valuable medical decision markers for coronary artery disease. Computational fluid dynamics (CFD) is the gold standard for accurate, non-invasive evaluation of these quantities in vivo. In this work, we propose a time-efficient surrogate model, powered by machine learning, for the estimation of pulsatile hemodynamics based on steady-state priors. We introduce deep vectorised operators, a modelling framework for discretisation independent learning on infinite-dimensional function spaces. The underlying neural architecture is a neural field conditioned on hemodynamic boundary conditions. Importantly, we show how relaxing the requirement of point-wise action to permutation-equivariance leads to a family of models that can be parametrised by message passing and self-attention layers. We evaluate our approach on a dataset of 74 stenotic coronary arteries extracted from coronary computed tomography angiography (CCTA) with patient-specific pulsatile CFD simulations as ground truth. We show that our model produces accurate estimates of the pulsatile velocity and pressure while being agnostic to re-sampling of the source domain (discretisation independence). This shows that deep vectorised operators are a powerful modelling tool for cardiovascular hemodynamics estimation in coronary arteries and beyond.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge