Bram de Wilde
Beyond Pixels: Medical Image Quality Assessment with Implicit Neural Representations
Aug 07, 2025Abstract:Artifacts pose a significant challenge in medical imaging, impacting diagnostic accuracy and downstream analysis. While image-based approaches for detecting artifacts can be effective, they often rely on preprocessing methods that can lead to information loss and high-memory-demand medical images, thereby limiting the scalability of classification models. In this work, we propose the use of implicit neural representations (INRs) for image quality assessment. INRs provide a compact and continuous representation of medical images, naturally handling variations in resolution and image size while reducing memory overhead. We develop deep weight space networks, graph neural networks, and relational attention transformers that operate on INRs to achieve image quality assessment. Our method is evaluated on the ACDC dataset with synthetically generated artifact patterns, demonstrating its effectiveness in assessing image quality while achieving similar performance with fewer parameters.
Steerable Anatomical Shape Synthesis with Implicit Neural Representations
Apr 04, 2025
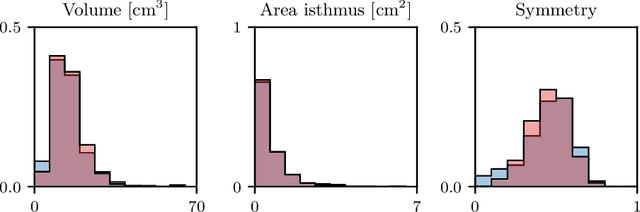
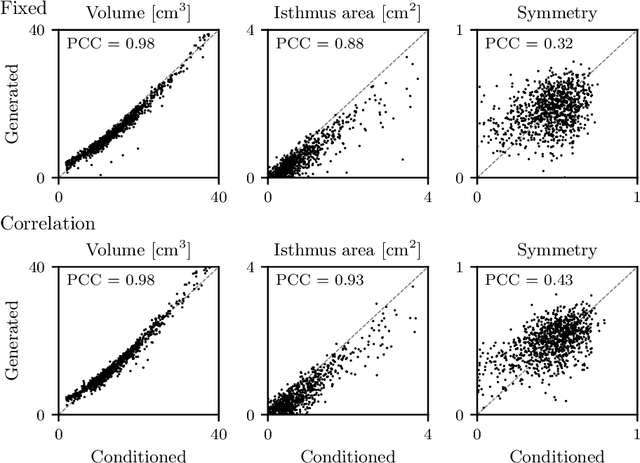
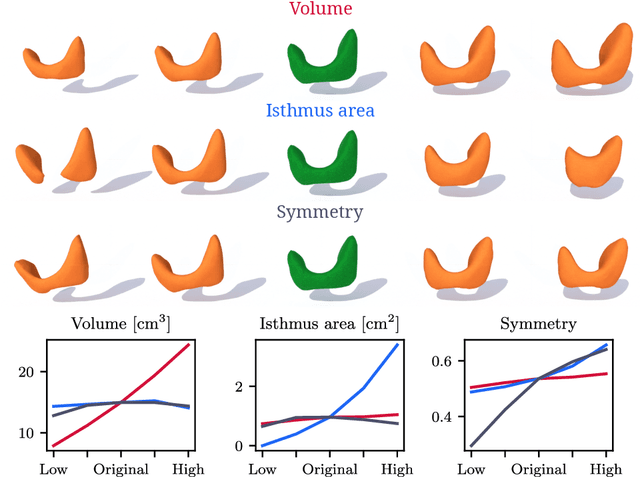
Abstract:Generative modeling of anatomical structures plays a crucial role in virtual imaging trials, which allow researchers to perform studies without the costs and constraints inherent to in vivo and phantom studies. For clinical relevance, generative models should allow targeted control to simulate specific patient populations rather than relying on purely random sampling. In this work, we propose a steerable generative model based on implicit neural representations. Implicit neural representations naturally support topology changes, making them well-suited for anatomical structures with varying topology, such as the thyroid. Our model learns a disentangled latent representation, enabling fine-grained control over shape variations. Evaluation includes reconstruction accuracy and anatomical plausibility. Our results demonstrate that the proposed model achieves high-quality shape generation while enabling targeted anatomical modifications.
Unleashing the Strengths of Unlabeled Data in Pan-cancer Abdominal Organ Quantification: the FLARE22 Challenge
Aug 10, 2023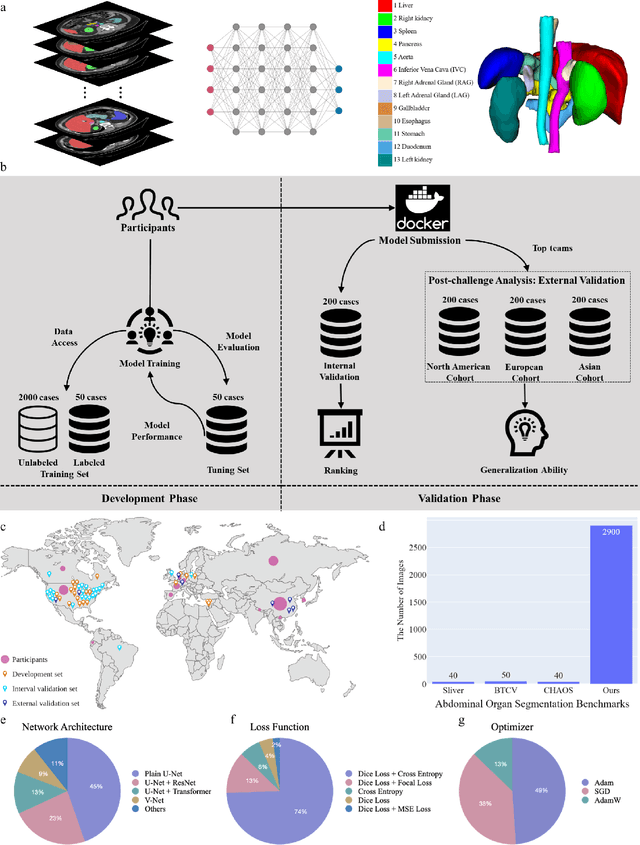
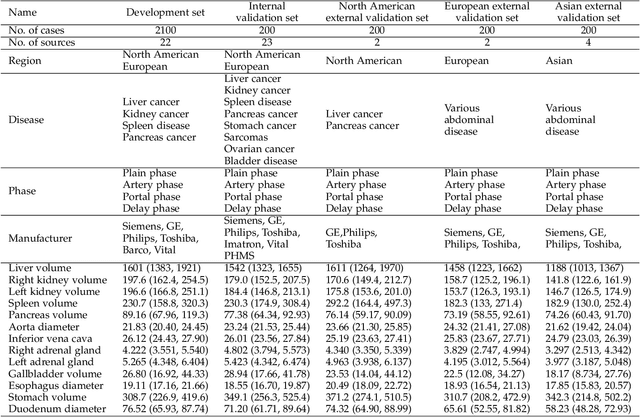
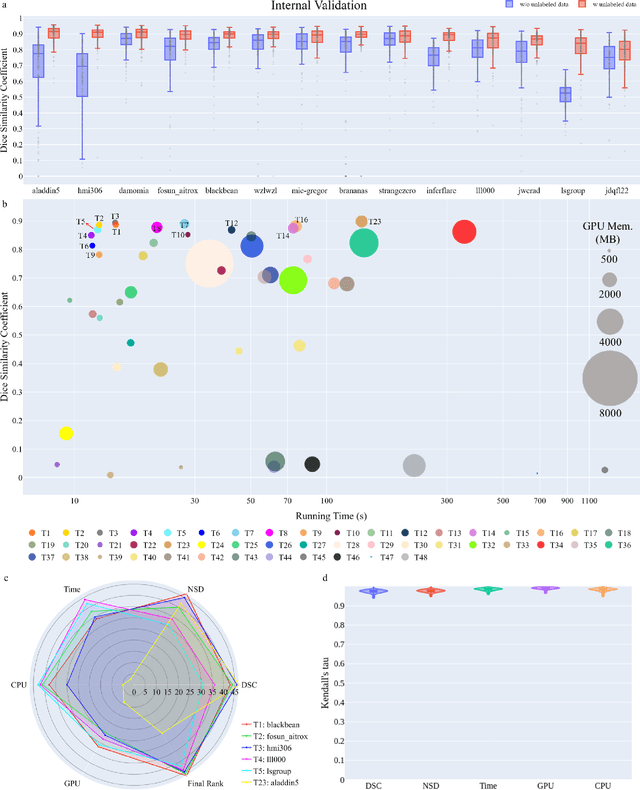
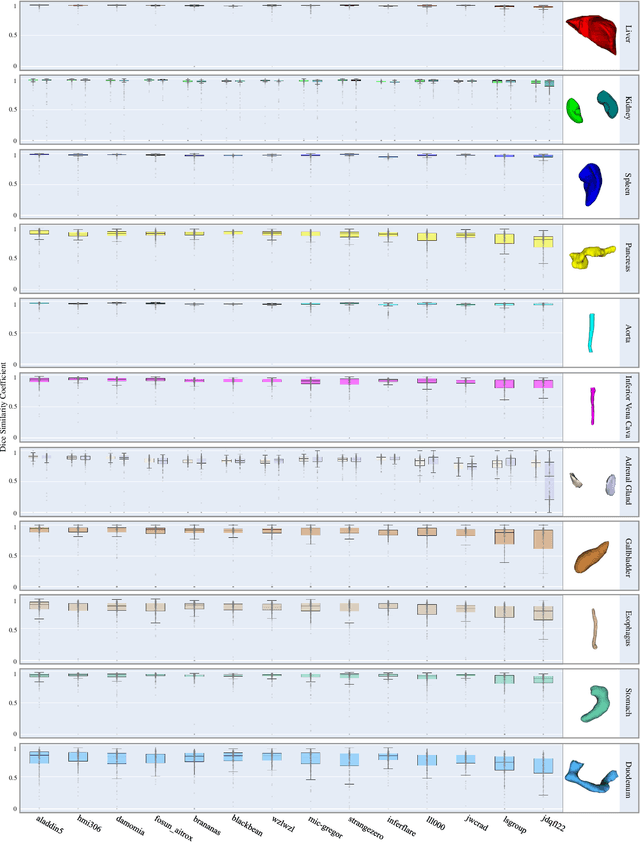
Abstract:Quantitative organ assessment is an essential step in automated abdominal disease diagnosis and treatment planning. Artificial intelligence (AI) has shown great potential to automatize this process. However, most existing AI algorithms rely on many expert annotations and lack a comprehensive evaluation of accuracy and efficiency in real-world multinational settings. To overcome these limitations, we organized the FLARE 2022 Challenge, the largest abdominal organ analysis challenge to date, to benchmark fast, low-resource, accurate, annotation-efficient, and generalized AI algorithms. We constructed an intercontinental and multinational dataset from more than 50 medical groups, including Computed Tomography (CT) scans with different races, diseases, phases, and manufacturers. We independently validated that a set of AI algorithms achieved a median Dice Similarity Coefficient (DSC) of 90.0\% by using 50 labeled scans and 2000 unlabeled scans, which can significantly reduce annotation requirements. The best-performing algorithms successfully generalized to holdout external validation sets, achieving a median DSC of 89.5\%, 90.9\%, and 88.3\% on North American, European, and Asian cohorts, respectively. They also enabled automatic extraction of key organ biology features, which was labor-intensive with traditional manual measurements. This opens the potential to use unlabeled data to boost performance and alleviate annotation shortages for modern AI models.
Medical diffusion on a budget: textual inversion for medical image generation
Mar 23, 2023



Abstract:Diffusion-based models for text-to-image generation have gained immense popularity due to recent advancements in efficiency, accessibility, and quality. Although it is becoming increasingly feasible to perform inference with these systems using consumer-grade GPUs, training them from scratch still requires access to large datasets and significant computational resources. In the case of medical image generation, the availability of large, publicly accessible datasets that include text reports is limited due to legal and ethical concerns. While training a diffusion model on a private dataset may address this issue, it is not always feasible for institutions lacking the necessary computational resources. This work demonstrates that pre-trained Stable Diffusion models, originally trained on natural images, can be adapted to various medical imaging modalities by training text embeddings with textual inversion. In this study, we conducted experiments using medical datasets comprising only 100 samples from three medical modalities. Embeddings were trained in a matter of hours, while still retaining diagnostic relevance in image generation. Experiments were designed to achieve several objectives. Firstly, we fine-tuned the training and inference processes of textual inversion, revealing that larger embeddings and more examples are required. Secondly, we validated our approach by demonstrating a 2\% increase in the diagnostic accuracy (AUC) for detecting prostate cancer on MRI, which is a challenging multi-modal imaging modality, from 0.78 to 0.80. Thirdly, we performed simulations by interpolating between healthy and diseased states, combining multiple pathologies, and inpainting to show embedding flexibility and control of disease appearance. Finally, the embeddings trained in this study are small (less than 1 MB), which facilitates easy sharing of medical data with reduced privacy concerns.
Cine-MRI detection of abdominal adhesions with spatio-temporal deep learning
Jun 15, 2021
Abstract:Adhesions are an important cause of chronic pain following abdominal surgery. Recent developments in abdominal cine-MRI have enabled the non-invasive diagnosis of adhesions. Adhesions are identified on cine-MRI by the absence of sliding motion during movement. Diagnosis and mapping of adhesions improves the management of patients with pain. Detection of abdominal adhesions on cine-MRI is challenging from both a radiological and deep learning perspective. We focus on classifying presence or absence of adhesions in sagittal abdominal cine-MRI series. We experimented with spatio-temporal deep learning architectures centered around a ConvGRU architecture. A hybrid architecture comprising a ResNet followed by a ConvGRU model allows to classify a whole time-series. Compared to a stand-alone ResNet with a two time-point (inspiration/expiration) input, we show an increase in classification performance (AUROC) from 0.74 to 0.83 ($p<0.05$). Our full temporal classification approach adds only a small amount (5%) of parameters to the entire architecture, which may be useful for other medical imaging problems with a temporal dimension.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge