Song Gu
Automatic Organ and Pan-cancer Segmentation in Abdomen CT: the FLARE 2023 Challenge
Aug 22, 2024Abstract:Organ and cancer segmentation in abdomen Computed Tomography (CT) scans is the prerequisite for precise cancer diagnosis and treatment. Most existing benchmarks and algorithms are tailored to specific cancer types, limiting their ability to provide comprehensive cancer analysis. This work presents the first international competition on abdominal organ and pan-cancer segmentation by providing a large-scale and diverse dataset, including 4650 CT scans with various cancer types from over 40 medical centers. The winning team established a new state-of-the-art with a deep learning-based cascaded framework, achieving average Dice Similarity Coefficient scores of 92.3% for organs and 64.9% for lesions on the hidden multi-national testing set. The dataset and code of top teams are publicly available, offering a benchmark platform to drive further innovations https://codalab.lisn.upsaclay.fr/competitions/12239.
The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions
Aug 10, 2023Abstract:Cell segmentation is a critical step for quantitative single-cell analysis in microscopy images. Existing cell segmentation methods are often tailored to specific modalities or require manual interventions to specify hyperparameters in different experimental settings. Here, we present a multi-modality cell segmentation benchmark, comprising over 1500 labeled images derived from more than 50 diverse biological experiments. The top participants developed a Transformer-based deep-learning algorithm that not only exceeds existing methods, but can also be applied to diverse microscopy images across imaging platforms and tissue types without manual parameter adjustments. This benchmark and the improved algorithm offer promising avenues for more accurate and versatile cell analysis in microscopy imaging.
Unleashing the Strengths of Unlabeled Data in Pan-cancer Abdominal Organ Quantification: the FLARE22 Challenge
Aug 10, 2023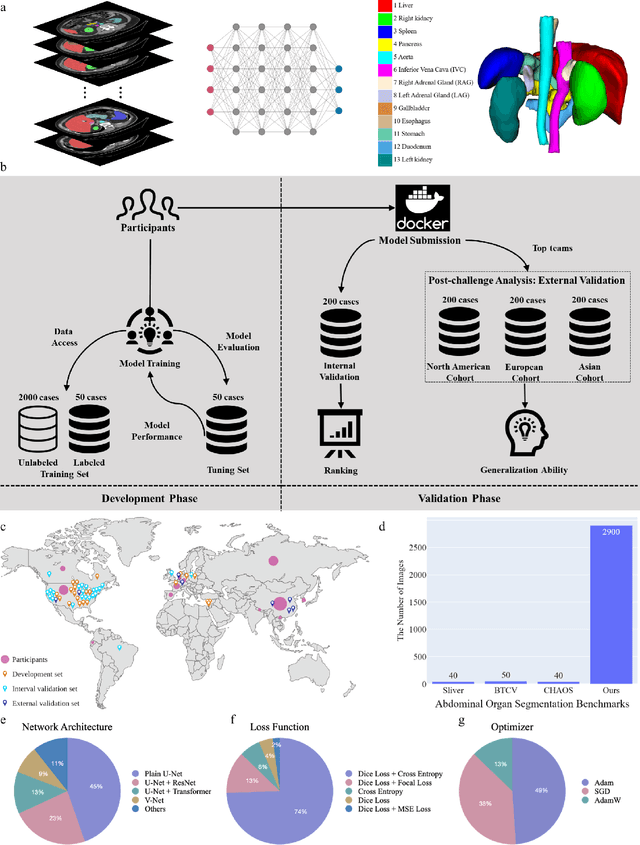
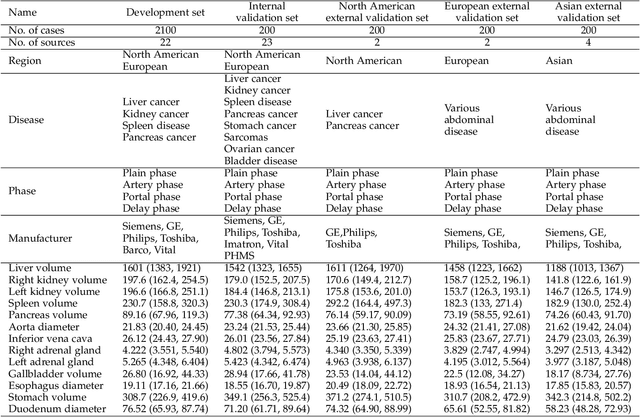
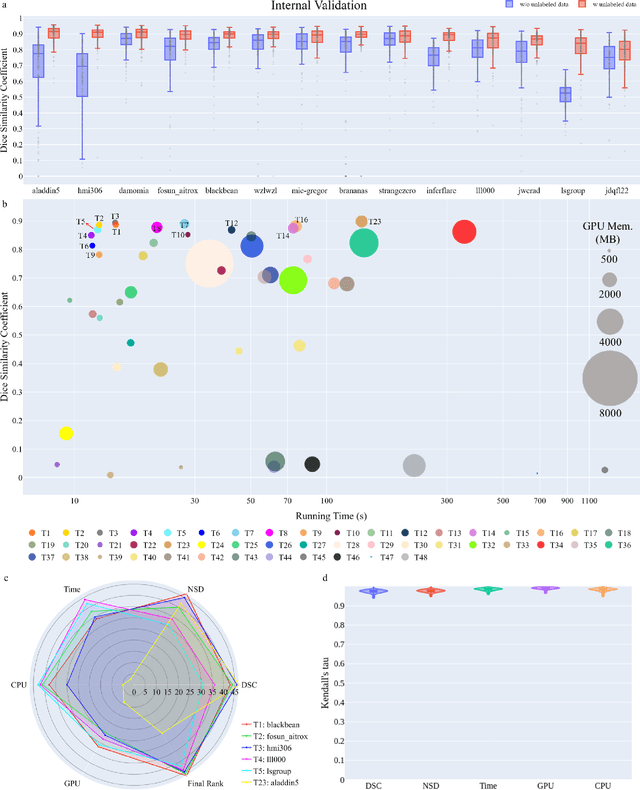
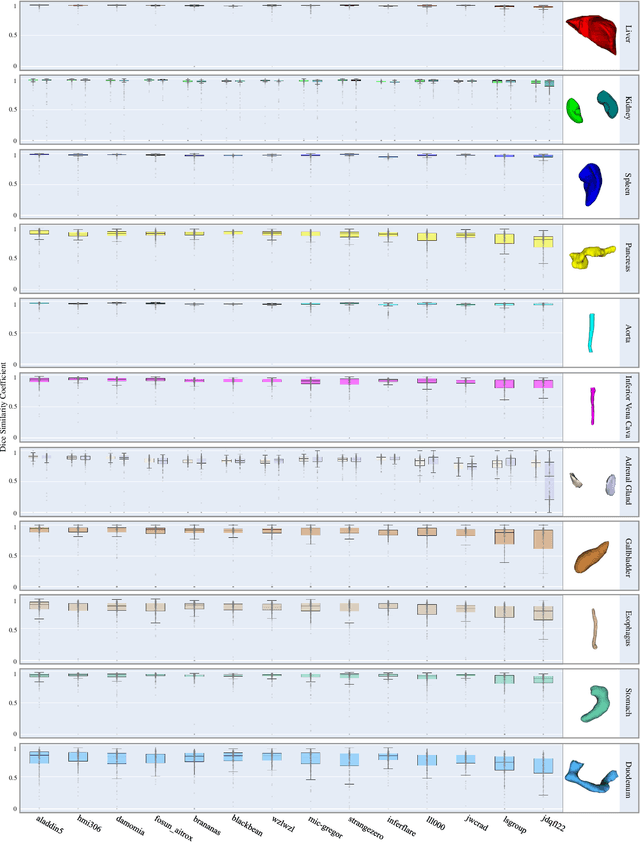
Abstract:Quantitative organ assessment is an essential step in automated abdominal disease diagnosis and treatment planning. Artificial intelligence (AI) has shown great potential to automatize this process. However, most existing AI algorithms rely on many expert annotations and lack a comprehensive evaluation of accuracy and efficiency in real-world multinational settings. To overcome these limitations, we organized the FLARE 2022 Challenge, the largest abdominal organ analysis challenge to date, to benchmark fast, low-resource, accurate, annotation-efficient, and generalized AI algorithms. We constructed an intercontinental and multinational dataset from more than 50 medical groups, including Computed Tomography (CT) scans with different races, diseases, phases, and manufacturers. We independently validated that a set of AI algorithms achieved a median Dice Similarity Coefficient (DSC) of 90.0\% by using 50 labeled scans and 2000 unlabeled scans, which can significantly reduce annotation requirements. The best-performing algorithms successfully generalized to holdout external validation sets, achieving a median DSC of 89.5\%, 90.9\%, and 88.3\% on North American, European, and Asian cohorts, respectively. They also enabled automatic extraction of key organ biology features, which was labor-intensive with traditional manual measurements. This opens the potential to use unlabeled data to boost performance and alleviate annotation shortages for modern AI models.
AbdomenCT-1K: Is Abdominal Organ Segmentation A Solved Problem?
Oct 28, 2020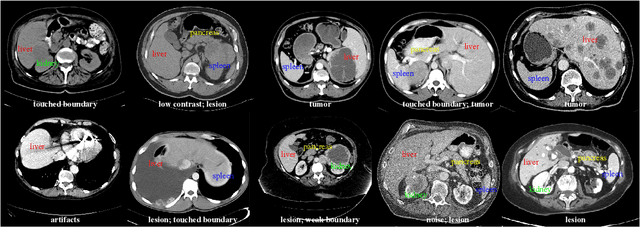
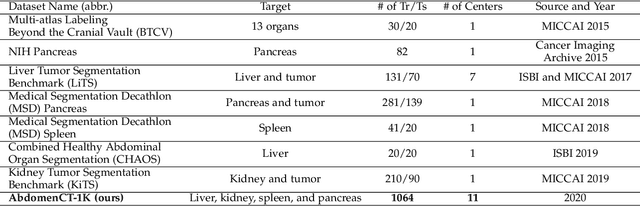
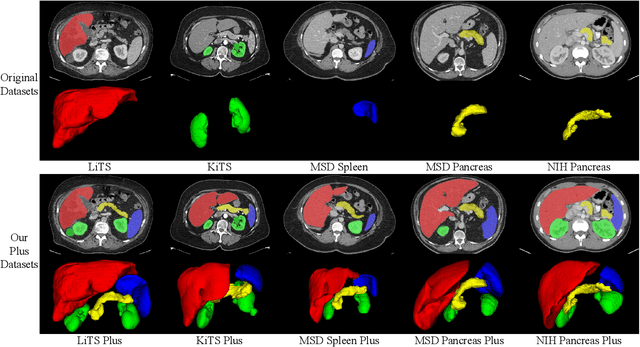
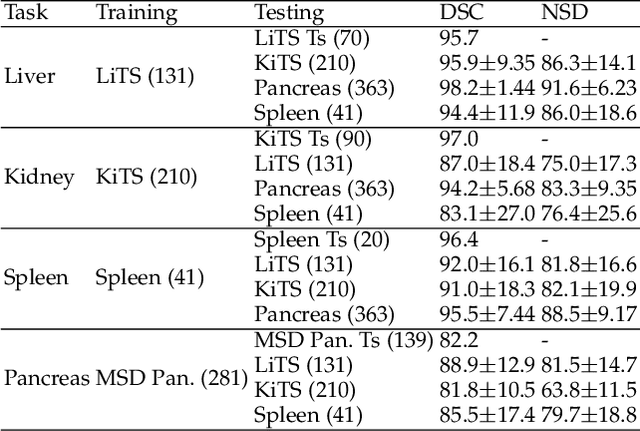
Abstract:With the unprecedented developments in deep learning, automatic segmentation of main abdominal organs (i.e., liver, kidney, and spleen) seems to be a solved problem as the state-of-the-art (SOTA) methods have achieved comparable results with inter-observer variability on existing benchmark datasets. However, most of the existing abdominal organ segmentation benchmark datasets only contain single-center, single-phase, single-vendor, or single-disease cases, thus, it is unclear whether the excellent performance can generalize on more diverse datasets. In this paper, we present a large and diverse abdominal CT organ segmentation dataset, termed as AbdomenCT-1K, with more than 1000 (1K) CT scans from 11 countries, including multi-center, multi-phase, multi-vendor, and multi-disease cases. Furthermore, we conduct a large-scale study for liver, kidney, spleen, and pancreas segmentation, as well as reveal the unsolved segmentation problems of the SOTA method, such as the limited generalization ability on distinct medical centers, phases, and unseen diseases. To advance the unsolved problems, we build four organ segmentation benchmarks for fully supervised, semi-supervised, weakly supervised, and continual learning, which are currently challenging and active research topics. Accordingly, we develop a simple and effective method for each benchmark, which can be used as out-of-the-box methods and strong baselines. We believe the introduction of the AbdomenCT-1K dataset will promote future in-depth research towards clinical applicable abdominal organ segmentation methods. Moreover, the datasets, codes, and trained models of baseline methods will be publicly available at https://github.com/JunMa11/AbdomenCT-1K.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge