Ritu Gupta
The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions
Aug 10, 2023Abstract:Cell segmentation is a critical step for quantitative single-cell analysis in microscopy images. Existing cell segmentation methods are often tailored to specific modalities or require manual interventions to specify hyperparameters in different experimental settings. Here, we present a multi-modality cell segmentation benchmark, comprising over 1500 labeled images derived from more than 50 diverse biological experiments. The top participants developed a Transformer-based deep-learning algorithm that not only exceeds existing methods, but can also be applied to diverse microscopy images across imaging platforms and tissue types without manual parameter adjustments. This benchmark and the improved algorithm offer promising avenues for more accurate and versatile cell analysis in microscopy imaging.
SDCT-AuxNet$^θ$: DCT Augmented Stain Deconvolutional CNN with Auxiliary Classifier for Cancer Diagnosis
Jun 08, 2020

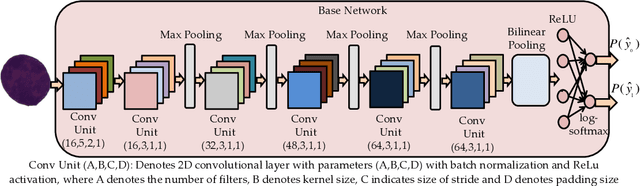
Abstract:Acute lymphoblastic leukemia (ALL) is a pervasive pediatric white blood cell cancer across the globe. With the popularity of convolutional neural networks (CNNs), computer-aided diagnosis of cancer has attracted considerable attention. Such tools are easily deployable and are cost-effective. Hence, these can enable extensive coverage of cancer diagnostic facilities. However, the development of such a tool for ALL cancer was challenging so far due to the non-availability of a large training dataset. The visual similarity between the malignant and normal cells adds to the complexity of the problem. This paper discusses the recent release of a large dataset and presents a novel deep learning architecture for the classification of cell images of ALL cancer. The proposed architecture, namely, SDCT-AuxNet$^{\theta}$ is a 2-module framework that utilizes a compact CNN as the main classifier in one module and a Kernel SVM as the auxiliary classifier in the other one. While CNN classifier uses features through bilinear-pooling, spectral-averaged features are used by the auxiliary classifier. Further, this CNN is trained on the stain deconvolved quantity images in the optical density domain instead of the conventional RGB images. A novel test strategy is proposed that exploits both the classifiers for decision making using the confidence scores of their predicted class labels. Elaborate experiments have been carried out on our recently released public dataset of 15114 images of ALL cancer and healthy cells to establish the validity of the proposed methodology that is also robust to subject-level variability. A weighted F1 score of 94.8$\%$ is obtained that is best so far on this challenging dataset.
* The final version of this preprint has been published in Medical Image Analysis
Heterogeneity Loss to Handle Intersubject and Intrasubject Variability in Cancer
Mar 19, 2020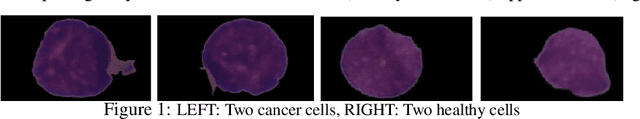
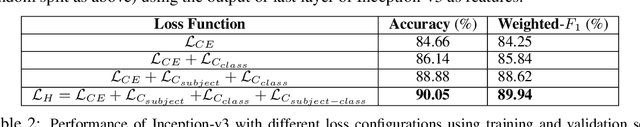
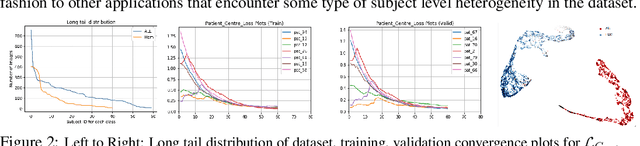
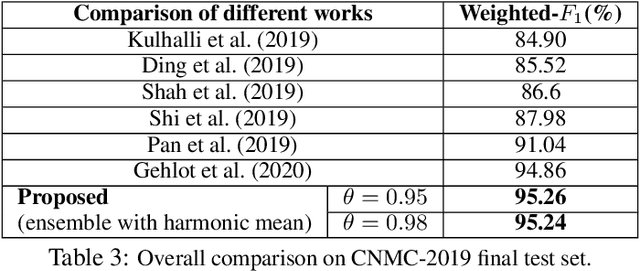
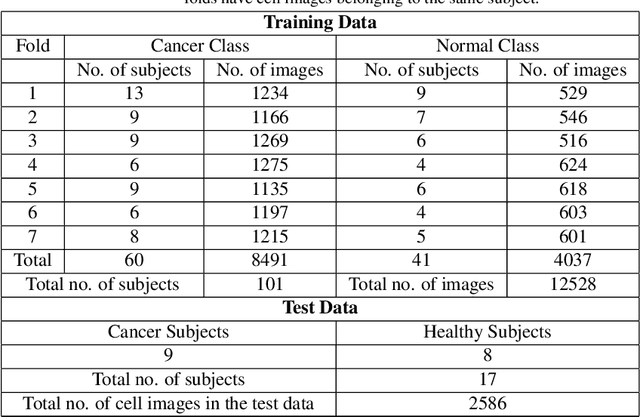
Abstract:Developing nations lack adequate number of hospitals with modern equipment and skilled doctors. Hence, a significant proportion of these nations' population, particularly in rural areas, is not able to avail specialized and timely healthcare facilities. In recent years, deep learning (DL) models, a class of artificial intelligence (AI) methods, have shown impressive results in medical domain. These AI methods can provide immense support to developing nations as affordable healthcare solutions. This work is focused on one such application of blood cancer diagnosis. However, there are some challenges to DL models in cancer research because of the unavailability of a large data for adequate training and the difficulty of capturing heterogeneity in data at different levels ranging from acquisition characteristics, session, to subject-level (within subjects and across subjects). These challenges render DL models prone to overfitting and hence, models lack generalization on prospective subjects' data. In this work, we address these problems in the application of B-cell Acute Lymphoblastic Leukemia (B-ALL) diagnosis using deep learning. We propose heterogeneity loss that captures subject-level heterogeneity, thereby, forcing the neural network to learn subject-independent features. We also propose an unorthodox ensemble strategy that helps us in providing improved classification over models trained on 7-folds giving a weighted-$F_1$ score of 95.26% on unseen (test) subjects' data that are, so far, the best results on the C-NMC 2019 dataset for B-ALL classification.
LeukoNet: DCT-based CNN architecture for the classification of normal versus Leukemic blasts in B-ALL Cancer
Nov 04, 2018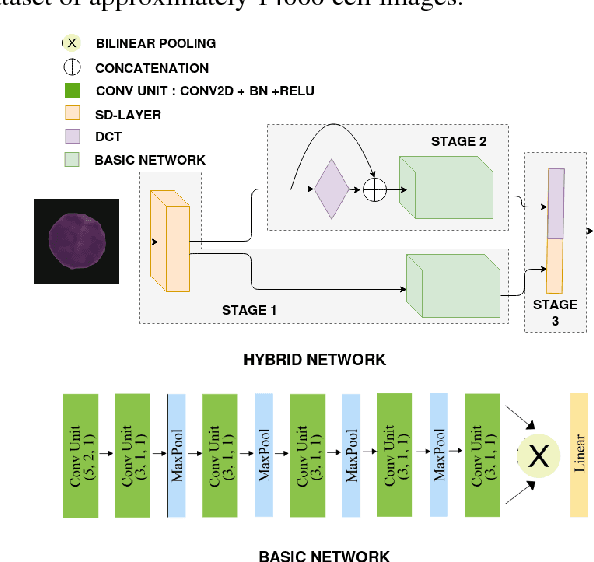
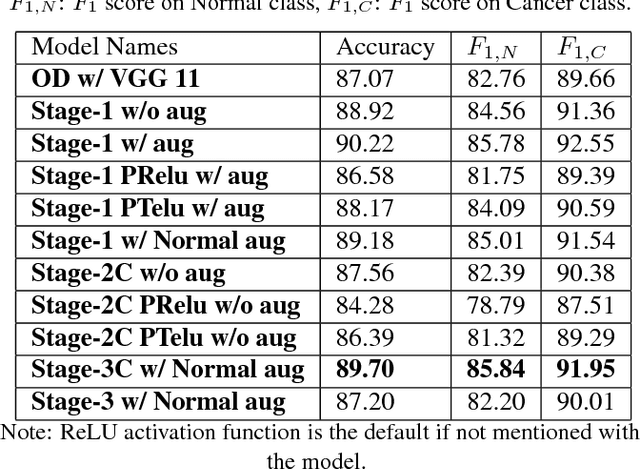
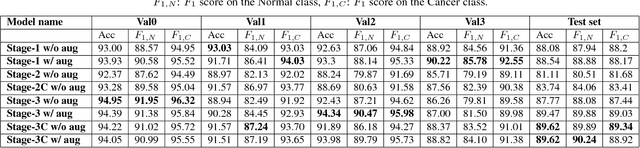
Abstract:Acute lymphoblastic leukemia (ALL) constitutes approximately 25% of the pediatric cancers. In general, the task of identifying immature leukemic blasts from normal cells under the microscope is challenging because morphologically the images of the two cells appear similar. In this paper, we propose a deep learning framework for classifying immature leukemic blasts and normal cells. The proposed model combines the Discrete Cosine Transform (DCT) domain features extracted via CNN with the Optical Density (OD) space features to build a robust classifier. Elaborate experiments have been conducted to validate the proposed LeukoNet classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge