Ronald Xie
WangLab at MEDIQA-M3G 2024: Multimodal Medical Answer Generation using Large Language Models
Apr 22, 2024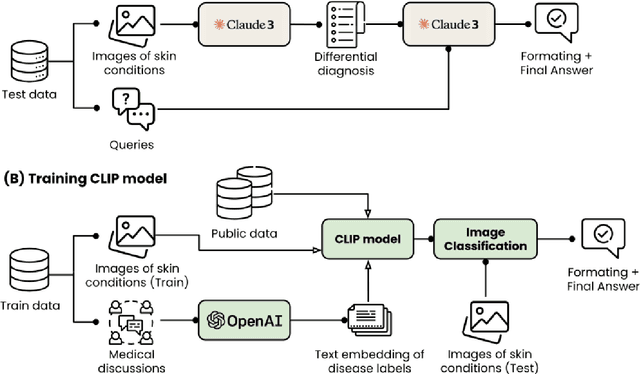
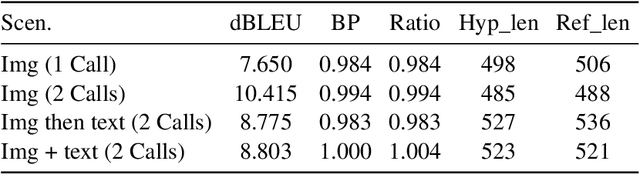
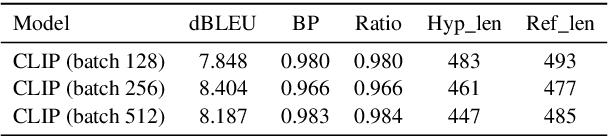
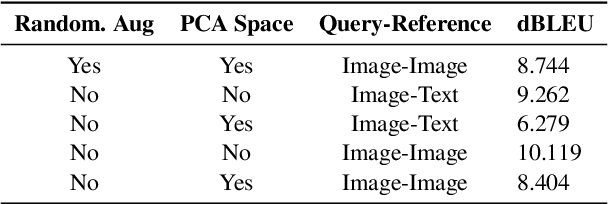
Abstract:This paper outlines our submission to the MEDIQA2024 Multilingual and Multimodal Medical Answer Generation (M3G) shared task. We report results for two standalone solutions under the English category of the task, the first involving two consecutive API calls to the Claude 3 Opus API and the second involving training an image-disease label joint embedding in the style of CLIP for image classification. These two solutions scored 1st and 2nd place respectively on the competition leaderboard, substantially outperforming the next best solution. Additionally, we discuss insights gained from post-competition experiments. While the performance of these two solutions have significant room for improvement due to the difficulty of the shared task and the challenging nature of medical visual question answering in general, we identify the multi-stage LLM approach and the CLIP image classification approach as promising avenues for further investigation.
WangLab at MEDIQA-CORR 2024: Optimized LLM-based Programs for Medical Error Detection and Correction
Apr 22, 2024



Abstract:Medical errors in clinical text pose significant risks to patient safety. The MEDIQA-CORR 2024 shared task focuses on detecting and correcting these errors across three subtasks: identifying the presence of an error, extracting the erroneous sentence, and generating a corrected sentence. In this paper, we present our approach that achieved top performance in all three subtasks. For the MS dataset, which contains subtle errors, we developed a retrieval-based system leveraging external medical question-answering datasets. For the UW dataset, reflecting more realistic clinical notes, we created a pipeline of modules to detect, localize, and correct errors. Both approaches utilized the DSPy framework for optimizing prompts and few-shot examples in large language model (LLM) based programs. Our results demonstrate the effectiveness of LLM based programs for medical error correction. However, our approach has limitations in addressing the full diversity of potential errors in medical documentation. We discuss the implications of our work and highlight future research directions to advance the robustness and applicability of medical error detection and correction systems.
The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions
Aug 10, 2023Abstract:Cell segmentation is a critical step for quantitative single-cell analysis in microscopy images. Existing cell segmentation methods are often tailored to specific modalities or require manual interventions to specify hyperparameters in different experimental settings. Here, we present a multi-modality cell segmentation benchmark, comprising over 1500 labeled images derived from more than 50 diverse biological experiments. The top participants developed a Transformer-based deep-learning algorithm that not only exceeds existing methods, but can also be applied to diverse microscopy images across imaging platforms and tissue types without manual parameter adjustments. This benchmark and the improved algorithm offer promising avenues for more accurate and versatile cell analysis in microscopy imaging.
Spatially Resolved Gene Expression Prediction from H&E Histology Images via Bi-modal Contrastive Learning
Jun 02, 2023



Abstract:Histology imaging is an important tool in medical diagnosis and research, enabling the examination of tissue structure and composition at the microscopic level. Understanding the underlying molecular mechanisms of tissue architecture is critical in uncovering disease mechanisms and developing effective treatments. Gene expression profiling provides insight into the molecular processes underlying tissue architecture, but the process can be time-consuming and expensive. In this study, we present BLEEP (Bi-modaL Embedding for Expression Prediction), a bi-modal embedding framework capable of generating spatially resolved gene expression profiles of whole-slide Hematoxylin and eosin (H&E) stained histology images. BLEEP uses a contrastive learning framework to construct a low-dimensional joint embedding space from a reference dataset using paired image and expression profiles at micrometer resolution. With this framework, the gene expression of any query image patch can be imputed using the expression profiles from the reference dataset. We demonstrate BLEEP's effectiveness in gene expression prediction by benchmarking its performance on a human liver tissue dataset captured via the 10x Visium platform, where it achieves significant improvements over existing methods. Our results demonstrate the potential of BLEEP to provide insights into the molecular mechanisms underlying tissue architecture, with important implications in diagnosis and research of various diseases. The proposed framework can significantly reduce the time and cost associated with gene expression profiling, opening up new avenues for high-throughput analysis of histology images for both research and clinical applications.
Clinical Note Generation from Doctor-Patient Conversations using Large Language Models: Insights from MEDIQA-Chat
May 03, 2023



Abstract:This paper describes our submission to the MEDIQA-Chat 2023 shared task for automatic clinical note generation from doctor-patient conversations. We report results for two approaches: the first fine-tunes a pre-trained language model (PLM) on the shared task data, and the second uses few-shot in-context learning (ICL) with a large language model (LLM). Both achieve high performance as measured by automatic metrics (e.g. ROUGE, BERTScore) and ranked second and first, respectively, of all submissions to the shared task. Expert human scrutiny indicates that notes generated via the ICL-based approach with GPT-4 are preferred about as often as human-written notes, making it a promising path toward automated note generation from doctor-patient conversations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge