Anubha Gupta
Exploring Multilingual Unseen Speaker Emotion Recognition: Leveraging Co-Attention Cues in Multitask Learning
Jun 13, 2024



Abstract:Advent of modern deep learning techniques has given rise to advancements in the field of Speech Emotion Recognition (SER). However, most systems prevalent in the field fail to generalize to speakers not seen during training. This study focuses on handling challenges of multilingual SER, specifically on unseen speakers. We introduce CAMuLeNet, a novel architecture leveraging co-attention based fusion and multitask learning to address this problem. Additionally, we benchmark pretrained encoders of Whisper, HuBERT, Wav2Vec2.0, and WavLM using 10-fold leave-speaker-out cross-validation on five existing multilingual benchmark datasets: IEMOCAP, RAVDESS, CREMA-D, EmoDB and CaFE and, release a novel dataset for SER on the Hindi language (BhavVani). CAMuLeNet shows an average improvement of approximately 8% over all benchmarks on unseen speakers determined by our cross-validation strategy.
CrossVoice: Crosslingual Prosody Preserving Cascade-S2ST using Transfer Learning
May 23, 2024


Abstract:This paper presents CrossVoice, a novel cascade-based Speech-to-Speech Translation (S2ST) system employing advanced ASR, MT, and TTS technologies with cross-lingual prosody preservation through transfer learning. We conducted comprehensive experiments comparing CrossVoice with direct-S2ST systems, showing improved BLEU scores on tasks such as Fisher Es-En, VoxPopuli Fr-En and prosody preservation on benchmark datasets CVSS-T and IndicTTS. With an average mean opinion score of 3.75 out of 4, speech synthesized by CrossVoice closely rivals human speech on the benchmark, highlighting the efficacy of cascade-based systems and transfer learning in multilingual S2ST with prosody transfer.
Multilingual Prosody Transfer: Comparing Supervised & Transfer Learning
May 23, 2024Abstract:The field of prosody transfer in speech synthesis systems is rapidly advancing. This research is focused on evaluating learning methods for adapting pre-trained monolingual text-to-speech (TTS) models to multilingual conditions, i.e., Supervised Fine-Tuning (SFT) and Transfer Learning (TL). This comparison utilizes three distinct metrics: Mean Opinion Score (MOS), Recognition Accuracy (RA), and Mel Cepstral Distortion (MCD). Results demonstrate that, in comparison to SFT, TL leads to significantly enhanced performance, with an average MOS higher by 1.53 points, a 37.5% increase in RA, and approximately a 7.8-point improvement in MCD. These findings are instrumental in helping build TTS models for low-resource languages.
The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions
Aug 10, 2023Abstract:Cell segmentation is a critical step for quantitative single-cell analysis in microscopy images. Existing cell segmentation methods are often tailored to specific modalities or require manual interventions to specify hyperparameters in different experimental settings. Here, we present a multi-modality cell segmentation benchmark, comprising over 1500 labeled images derived from more than 50 diverse biological experiments. The top participants developed a Transformer-based deep-learning algorithm that not only exceeds existing methods, but can also be applied to diverse microscopy images across imaging platforms and tissue types without manual parameter adjustments. This benchmark and the improved algorithm offer promising avenues for more accurate and versatile cell analysis in microscopy imaging.
Why is the winner the best?
Mar 30, 2023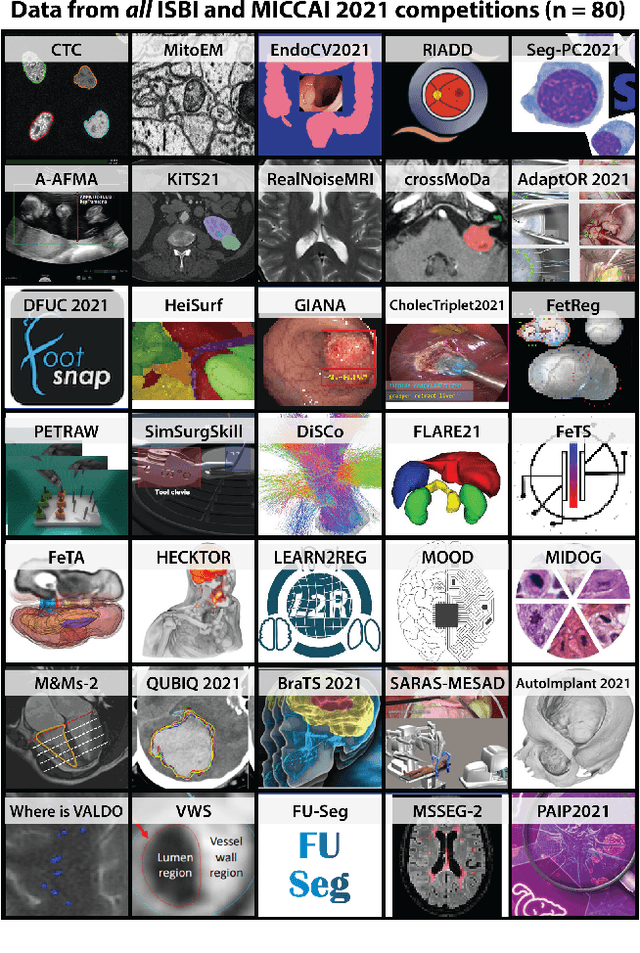
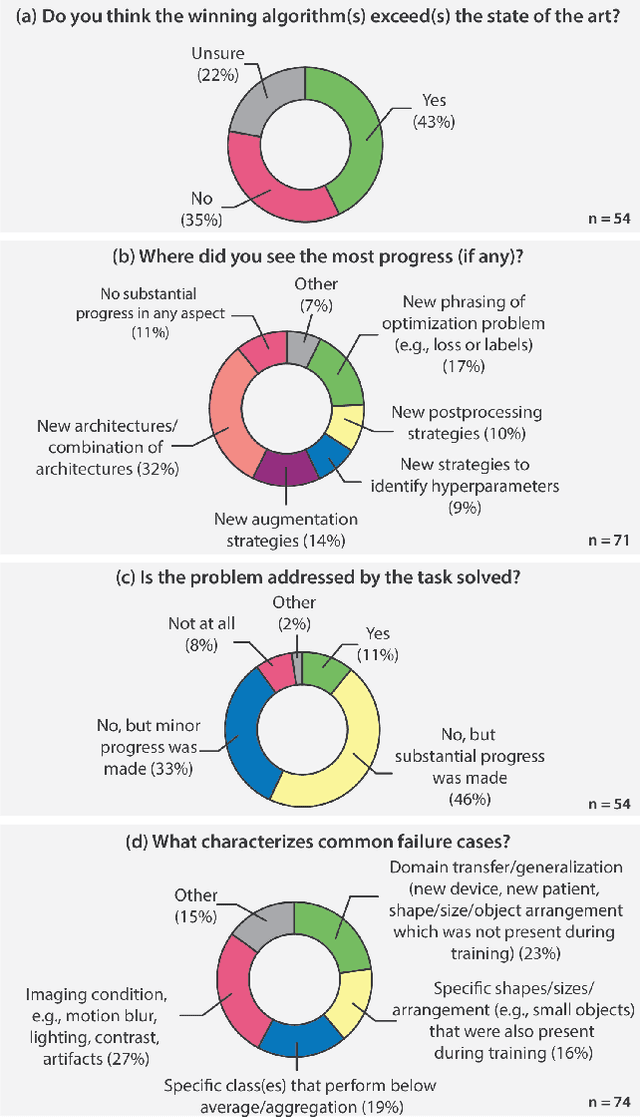
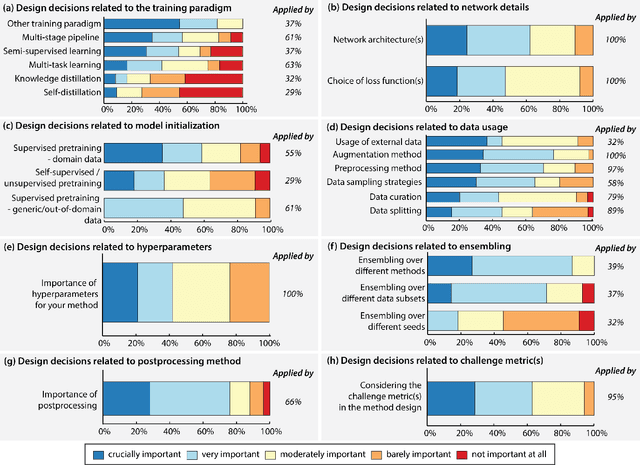
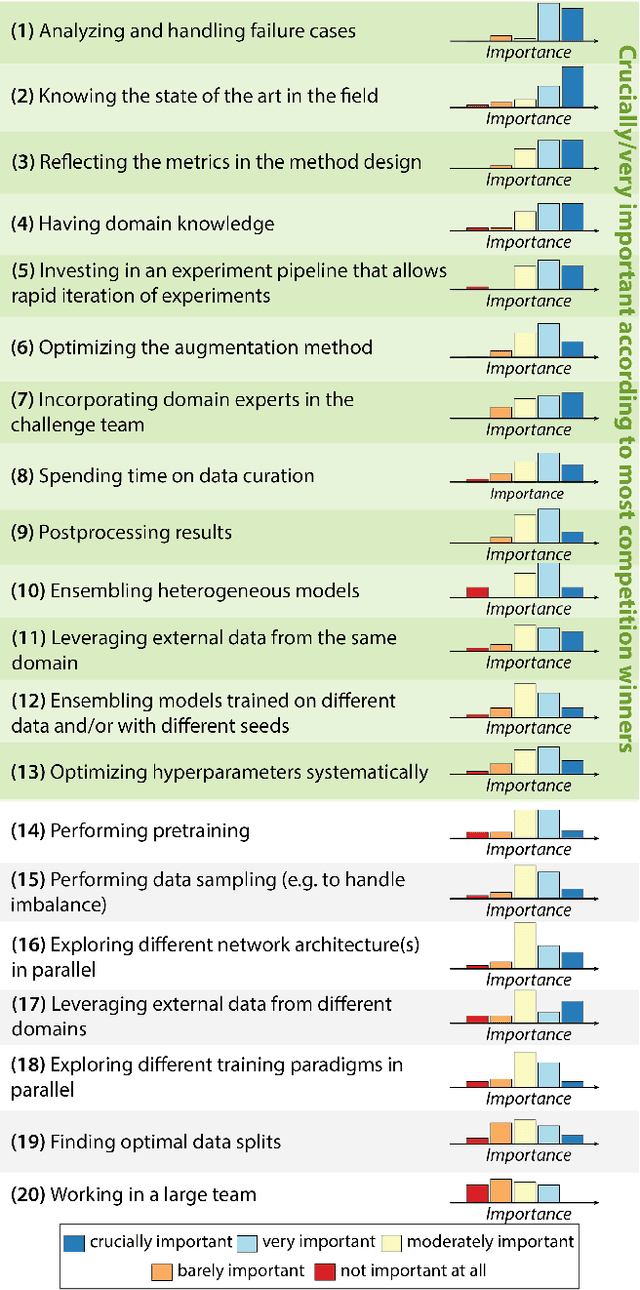
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
SDCT-AuxNet$^θ$: DCT Augmented Stain Deconvolutional CNN with Auxiliary Classifier for Cancer Diagnosis
Jun 08, 2020
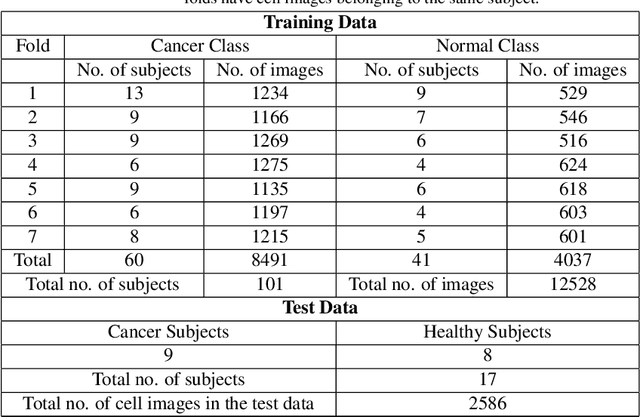

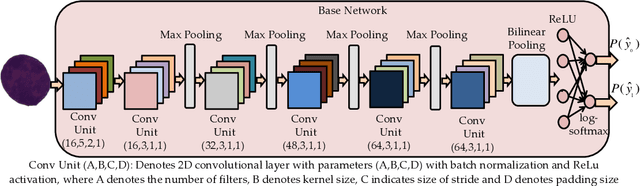
Abstract:Acute lymphoblastic leukemia (ALL) is a pervasive pediatric white blood cell cancer across the globe. With the popularity of convolutional neural networks (CNNs), computer-aided diagnosis of cancer has attracted considerable attention. Such tools are easily deployable and are cost-effective. Hence, these can enable extensive coverage of cancer diagnostic facilities. However, the development of such a tool for ALL cancer was challenging so far due to the non-availability of a large training dataset. The visual similarity between the malignant and normal cells adds to the complexity of the problem. This paper discusses the recent release of a large dataset and presents a novel deep learning architecture for the classification of cell images of ALL cancer. The proposed architecture, namely, SDCT-AuxNet$^{\theta}$ is a 2-module framework that utilizes a compact CNN as the main classifier in one module and a Kernel SVM as the auxiliary classifier in the other one. While CNN classifier uses features through bilinear-pooling, spectral-averaged features are used by the auxiliary classifier. Further, this CNN is trained on the stain deconvolved quantity images in the optical density domain instead of the conventional RGB images. A novel test strategy is proposed that exploits both the classifiers for decision making using the confidence scores of their predicted class labels. Elaborate experiments have been carried out on our recently released public dataset of 15114 images of ALL cancer and healthy cells to establish the validity of the proposed methodology that is also robust to subject-level variability. A weighted F1 score of 94.8$\%$ is obtained that is best so far on this challenging dataset.
* The final version of this preprint has been published in Medical Image Analysis
Heterogeneity Loss to Handle Intersubject and Intrasubject Variability in Cancer
Mar 19, 2020
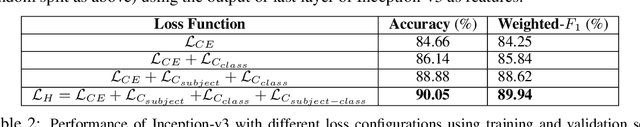
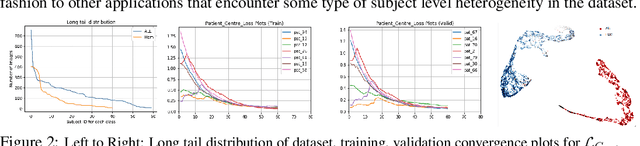
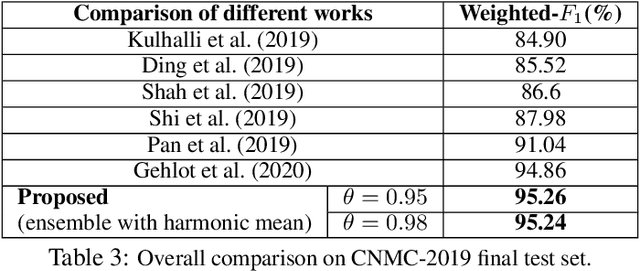
Abstract:Developing nations lack adequate number of hospitals with modern equipment and skilled doctors. Hence, a significant proportion of these nations' population, particularly in rural areas, is not able to avail specialized and timely healthcare facilities. In recent years, deep learning (DL) models, a class of artificial intelligence (AI) methods, have shown impressive results in medical domain. These AI methods can provide immense support to developing nations as affordable healthcare solutions. This work is focused on one such application of blood cancer diagnosis. However, there are some challenges to DL models in cancer research because of the unavailability of a large data for adequate training and the difficulty of capturing heterogeneity in data at different levels ranging from acquisition characteristics, session, to subject-level (within subjects and across subjects). These challenges render DL models prone to overfitting and hence, models lack generalization on prospective subjects' data. In this work, we address these problems in the application of B-cell Acute Lymphoblastic Leukemia (B-ALL) diagnosis using deep learning. We propose heterogeneity loss that captures subject-level heterogeneity, thereby, forcing the neural network to learn subject-independent features. We also propose an unorthodox ensemble strategy that helps us in providing improved classification over models trained on 7-folds giving a weighted-$F_1$ score of 95.26% on unseen (test) subjects' data that are, so far, the best results on the C-NMC 2019 dataset for B-ALL classification.
EDUQA: Educational Domain Question Answering System using Conceptual Network Mapping
Nov 12, 2019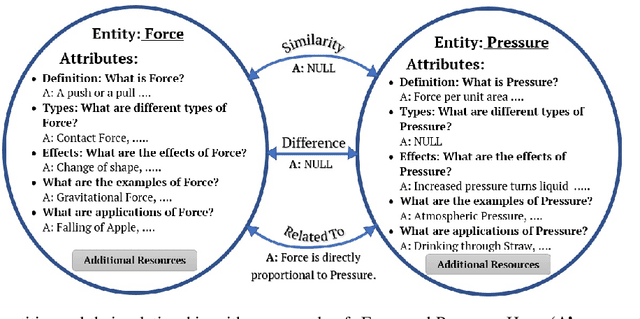
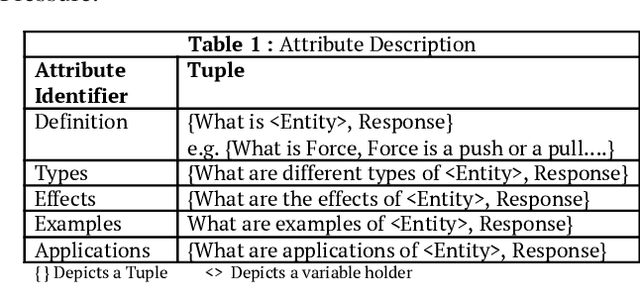
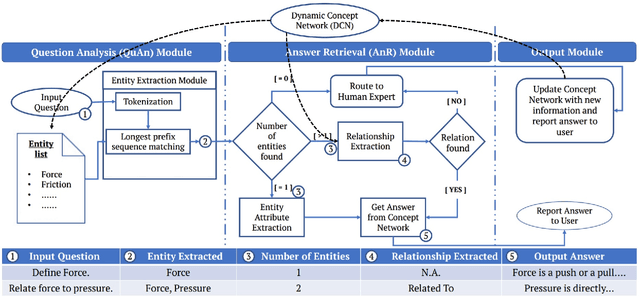
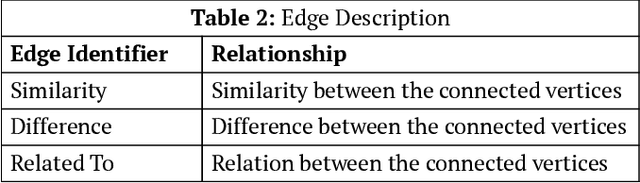
Abstract:Most of the existing question answering models can be largely compiled into two categories: i) open domain question answering models that answer generic questions and use large-scale knowledge base along with the targeted web-corpus retrieval and ii) closed domain question answering models that address focused questioning area and use complex deep learning models. Both the above models derive answers through textual comprehension methods. Due to their inability to capture the pedagogical meaning of textual content, these models are not appropriately suited to the educational field for pedagogy. In this paper, we propose an on-the-fly conceptual network model that incorporates educational semantics. The proposed model preserves correlations between conceptual entities by applying intelligent indexing algorithms on the concept network so as to improve answer generation. This model can be utilized for building interactive conversational agents for aiding classroom learning.
* Published in the 44th International Conference on Acoustics, Speech, and Signal Processing (ICASSP) 2019
LeukoNet: DCT-based CNN architecture for the classification of normal versus Leukemic blasts in B-ALL Cancer
Nov 04, 2018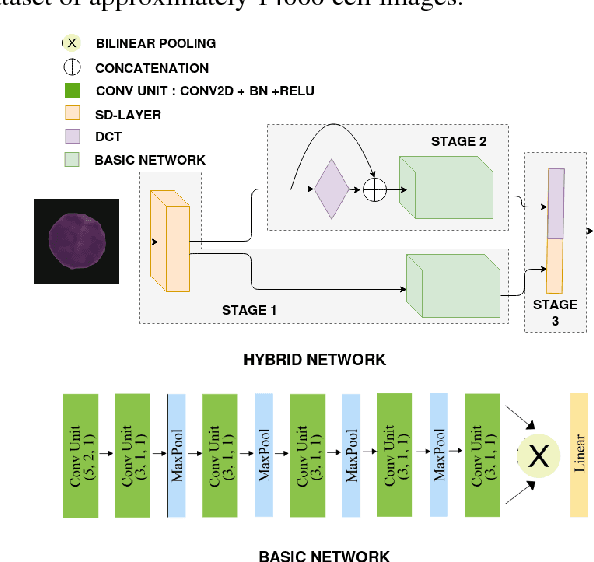
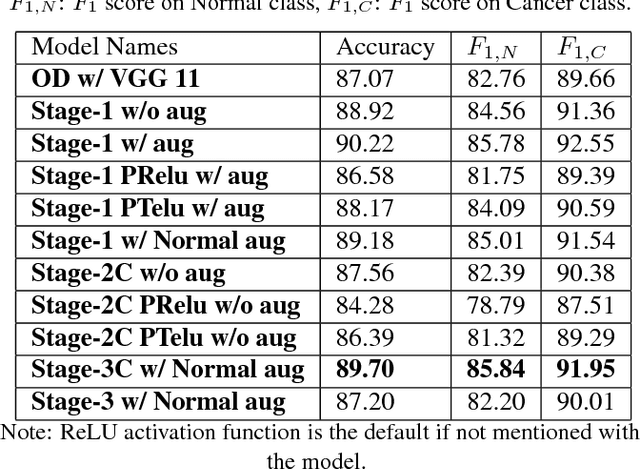
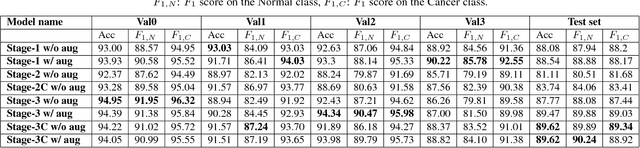
Abstract:Acute lymphoblastic leukemia (ALL) constitutes approximately 25% of the pediatric cancers. In general, the task of identifying immature leukemic blasts from normal cells under the microscope is challenging because morphologically the images of the two cells appear similar. In this paper, we propose a deep learning framework for classifying immature leukemic blasts and normal cells. The proposed model combines the Discrete Cosine Transform (DCT) domain features extracted via CNN with the Optical Density (OD) space features to build a robust classifier. Elaborate experiments have been conducted to validate the proposed LeukoNet classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge