Gary Bader
WangLab at MEDIQA-M3G 2024: Multimodal Medical Answer Generation using Large Language Models
Apr 22, 2024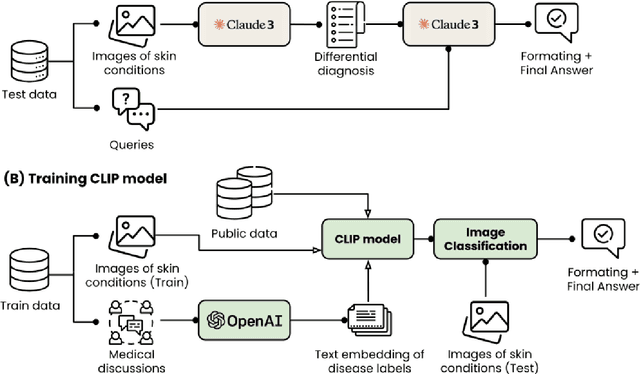
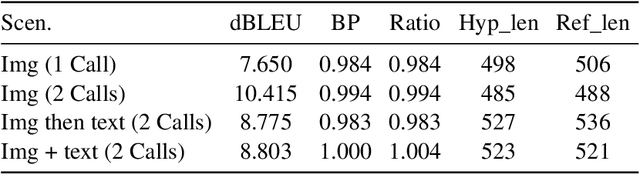
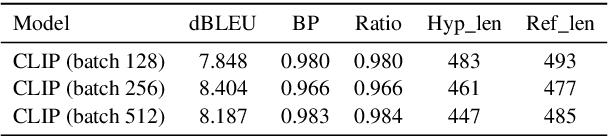
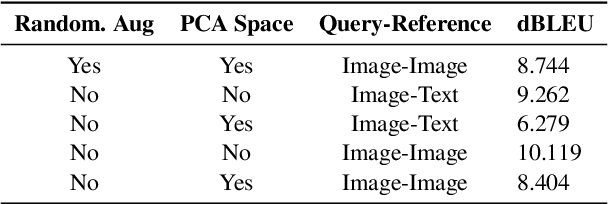
Abstract:This paper outlines our submission to the MEDIQA2024 Multilingual and Multimodal Medical Answer Generation (M3G) shared task. We report results for two standalone solutions under the English category of the task, the first involving two consecutive API calls to the Claude 3 Opus API and the second involving training an image-disease label joint embedding in the style of CLIP for image classification. These two solutions scored 1st and 2nd place respectively on the competition leaderboard, substantially outperforming the next best solution. Additionally, we discuss insights gained from post-competition experiments. While the performance of these two solutions have significant room for improvement due to the difficulty of the shared task and the challenging nature of medical visual question answering in general, we identify the multi-stage LLM approach and the CLIP image classification approach as promising avenues for further investigation.
Exploring the Challenges of Open Domain Multi-Document Summarization
Dec 20, 2022Abstract:Multi-document summarization (MDS) has traditionally been studied assuming a set of ground-truth topic-related input documents is provided. In practice, the input document set is unlikely to be available a priori and would need to be retrieved based on an information need, a setting we call open-domain MDS. We experiment with current state-of-the-art retrieval and summarization models on several popular MDS datasets extended to the open-domain setting. We find that existing summarizers suffer large reductions in performance when applied as-is to this more realistic task, though training summarizers with retrieved inputs can reduce their sensitivity retrieval errors. To further probe these findings, we conduct perturbation experiments on summarizer inputs to study the impact of different types of document retrieval errors. Based on our results, we provide practical guidelines to help facilitate a shift to open-domain MDS. We release our code and experimental results alongside all data or model artifacts created during our investigation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge