AbdomenCT-1K: Is Abdominal Organ Segmentation A Solved Problem?
Paper and Code
Oct 28, 2020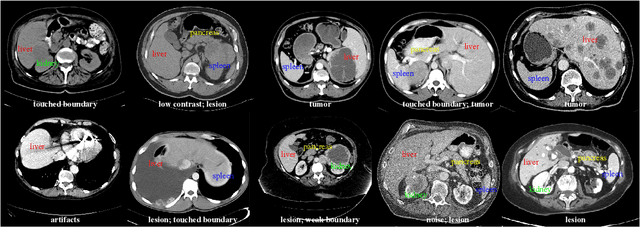
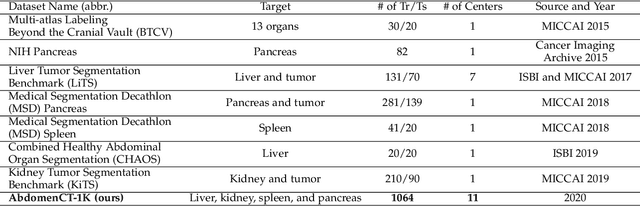
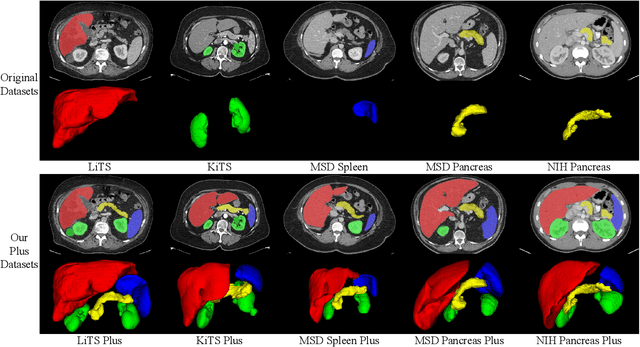
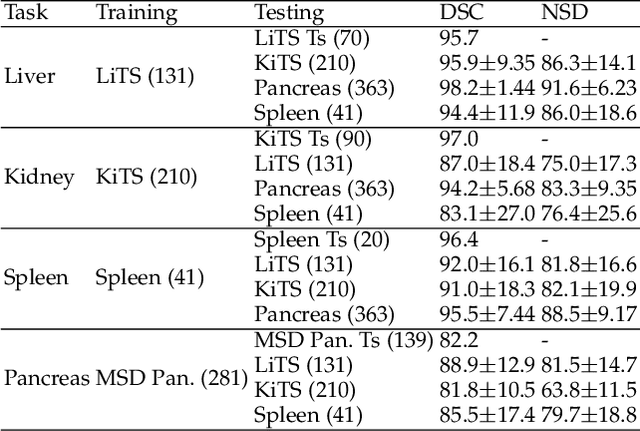
With the unprecedented developments in deep learning, automatic segmentation of main abdominal organs (i.e., liver, kidney, and spleen) seems to be a solved problem as the state-of-the-art (SOTA) methods have achieved comparable results with inter-observer variability on existing benchmark datasets. However, most of the existing abdominal organ segmentation benchmark datasets only contain single-center, single-phase, single-vendor, or single-disease cases, thus, it is unclear whether the excellent performance can generalize on more diverse datasets. In this paper, we present a large and diverse abdominal CT organ segmentation dataset, termed as AbdomenCT-1K, with more than 1000 (1K) CT scans from 11 countries, including multi-center, multi-phase, multi-vendor, and multi-disease cases. Furthermore, we conduct a large-scale study for liver, kidney, spleen, and pancreas segmentation, as well as reveal the unsolved segmentation problems of the SOTA method, such as the limited generalization ability on distinct medical centers, phases, and unseen diseases. To advance the unsolved problems, we build four organ segmentation benchmarks for fully supervised, semi-supervised, weakly supervised, and continual learning, which are currently challenging and active research topics. Accordingly, we develop a simple and effective method for each benchmark, which can be used as out-of-the-box methods and strong baselines. We believe the introduction of the AbdomenCT-1K dataset will promote future in-depth research towards clinical applicable abdominal organ segmentation methods. Moreover, the datasets, codes, and trained models of baseline methods will be publicly available at https://github.com/JunMa11/AbdomenCT-1K.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge