Cheng Zhu
MiniMax-M1: Scaling Test-Time Compute Efficiently with Lightning Attention
Jun 16, 2025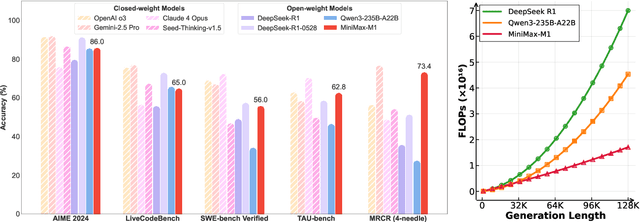
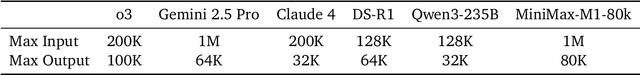
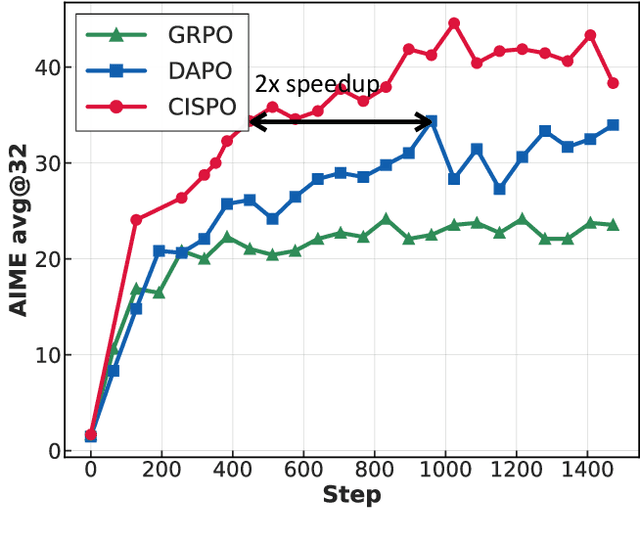
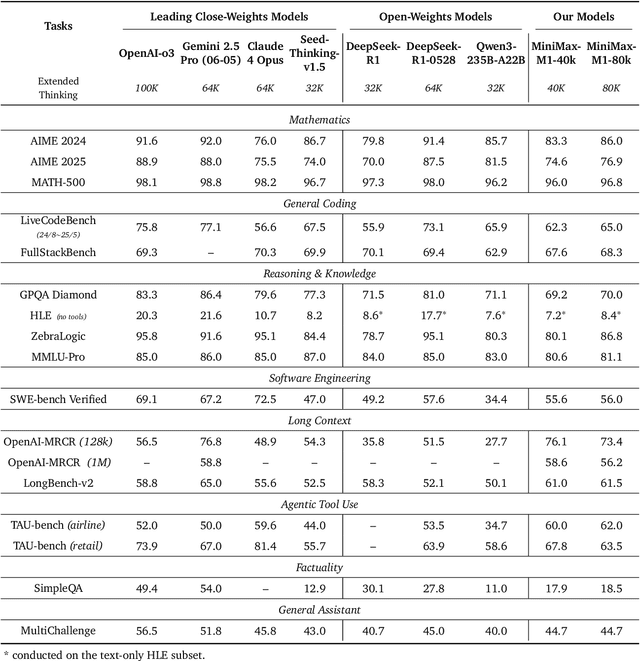
Abstract:We introduce MiniMax-M1, the world's first open-weight, large-scale hybrid-attention reasoning model. MiniMax-M1 is powered by a hybrid Mixture-of-Experts (MoE) architecture combined with a lightning attention mechanism. The model is developed based on our previous MiniMax-Text-01 model, which contains a total of 456 billion parameters with 45.9 billion parameters activated per token. The M1 model natively supports a context length of 1 million tokens, 8x the context size of DeepSeek R1. Furthermore, the lightning attention mechanism in MiniMax-M1 enables efficient scaling of test-time compute. These properties make M1 particularly suitable for complex tasks that require processing long inputs and thinking extensively. MiniMax-M1 is trained using large-scale reinforcement learning (RL) on diverse problems including sandbox-based, real-world software engineering environments. In addition to M1's inherent efficiency advantage for RL training, we propose CISPO, a novel RL algorithm to further enhance RL efficiency. CISPO clips importance sampling weights rather than token updates, outperforming other competitive RL variants. Combining hybrid-attention and CISPO enables MiniMax-M1's full RL training on 512 H800 GPUs to complete in only three weeks, with a rental cost of just $534,700. We release two versions of MiniMax-M1 models with 40K and 80K thinking budgets respectively, where the 40K model represents an intermediate phase of the 80K training. Experiments on standard benchmarks show that our models are comparable or superior to strong open-weight models such as the original DeepSeek-R1 and Qwen3-235B, with particular strengths in complex software engineering, tool utilization, and long-context tasks. We publicly release MiniMax-M1 at https://github.com/MiniMax-AI/MiniMax-M1.
Tool-as-Interface: Learning Robot Policies from Human Tool Usage through Imitation Learning
Apr 06, 2025Abstract:Tool use is critical for enabling robots to perform complex real-world tasks, and leveraging human tool-use data can be instrumental for teaching robots. However, existing data collection methods like teleoperation are slow, prone to control delays, and unsuitable for dynamic tasks. In contrast, human natural data, where humans directly perform tasks with tools, offers natural, unstructured interactions that are both efficient and easy to collect. Building on the insight that humans and robots can share the same tools, we propose a framework to transfer tool-use knowledge from human data to robots. Using two RGB cameras, our method generates 3D reconstruction, applies Gaussian splatting for novel view augmentation, employs segmentation models to extract embodiment-agnostic observations, and leverages task-space tool-action representations to train visuomotor policies. We validate our approach on diverse real-world tasks, including meatball scooping, pan flipping, wine bottle balancing, and other complex tasks. Our method achieves a 71\% higher average success rate compared to diffusion policies trained with teleoperation data and reduces data collection time by 77\%, with some tasks solvable only by our framework. Compared to hand-held gripper, our method cuts data collection time by 41\%. Additionally, our method bridges the embodiment gap, improves robustness to variations in camera viewpoints and robot configurations, and generalizes effectively across objects and spatial setups.
A Unified Modeling Framework for Automated Penetration Testing
Feb 17, 2025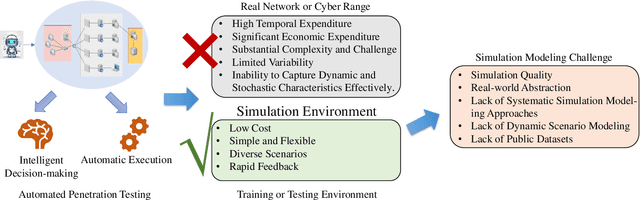
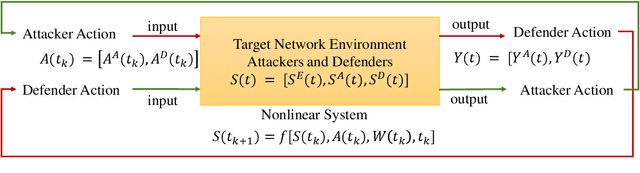
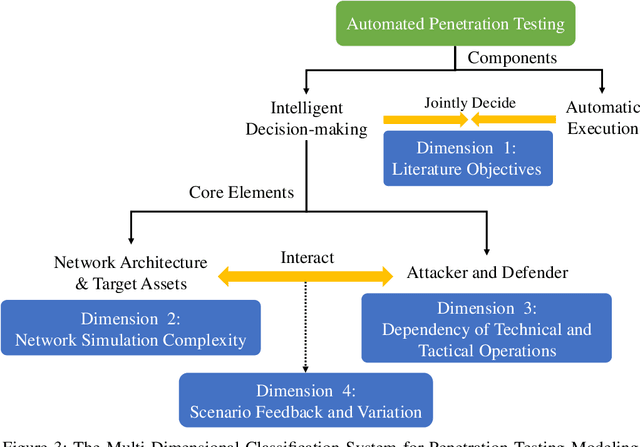
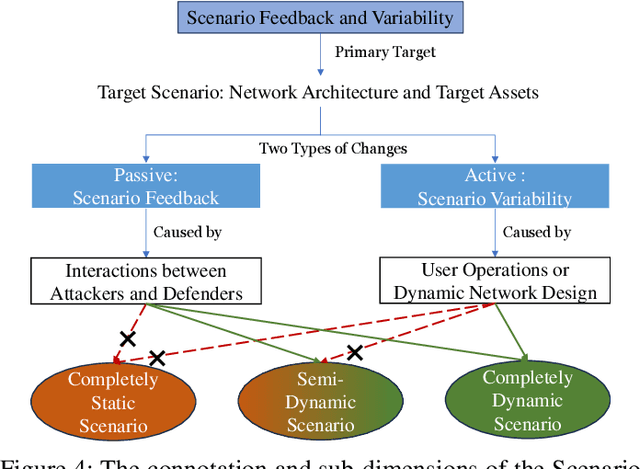
Abstract:The integration of artificial intelligence into automated penetration testing (AutoPT) has highlighted the necessity of simulation modeling for the training of intelligent agents, due to its cost-efficiency and swift feedback capabilities. Despite the proliferation of AutoPT research, there is a recognized gap in the availability of a unified framework for simulation modeling methods. This paper presents a systematic review and synthesis of existing techniques, introducing MDCPM to categorize studies based on literature objectives, network simulation complexity, dependency of technical and tactical operations, and scenario feedback and variation. To bridge the gap in unified method for multi-dimensional and multi-level simulation modeling, dynamic environment modeling, and the scarcity of public datasets, we introduce AutoPT-Sim, a novel modeling framework that based on policy automation and encompasses the combination of all sub dimensions. AutoPT-Sim offers a comprehensive approach to modeling network environments, attackers, and defenders, transcending the constraints of static modeling and accommodating networks of diverse scales. We publicly release a generated standard network environment dataset and the code of Network Generator. By integrating publicly available datasets flexibly, support is offered for various simulation modeling levels focused on policy automation in MDCPM and the network generator help researchers output customized target network data by adjusting parameters or fine-tuning the network generator.
MiniMax-01: Scaling Foundation Models with Lightning Attention
Jan 14, 2025Abstract:We introduce MiniMax-01 series, including MiniMax-Text-01 and MiniMax-VL-01, which are comparable to top-tier models while offering superior capabilities in processing longer contexts. The core lies in lightning attention and its efficient scaling. To maximize computational capacity, we integrate it with Mixture of Experts (MoE), creating a model with 32 experts and 456 billion total parameters, of which 45.9 billion are activated for each token. We develop an optimized parallel strategy and highly efficient computation-communication overlap techniques for MoE and lightning attention. This approach enables us to conduct efficient training and inference on models with hundreds of billions of parameters across contexts spanning millions of tokens. The context window of MiniMax-Text-01 can reach up to 1 million tokens during training and extrapolate to 4 million tokens during inference at an affordable cost. Our vision-language model, MiniMax-VL-01 is built through continued training with 512 billion vision-language tokens. Experiments on both standard and in-house benchmarks show that our models match the performance of state-of-the-art models like GPT-4o and Claude-3.5-Sonnet while offering 20-32 times longer context window. We publicly release MiniMax-01 at https://github.com/MiniMax-AI.
Edge-aware Hard Clustering Graph Pooling for Brain Imaging Data
Sep 13, 2023



Abstract:Graph Convolutional Networks (GCNs) can capture non-Euclidean spatial dependence between different brain regions, and the graph pooling operator in GCNs is key to enhancing the representation learning capability and acquiring abnormal brain maps. However, the majority of existing research designs graph pooling operators only from the perspective of nodes while disregarding the original edge features, in a way that not only confines graph pooling application scenarios, but also diminishes its ability to capture critical substructures. In this study, a clustering graph pooling method that first supports multidimensional edge features, called Edge-aware hard clustering graph pooling (EHCPool), is developed. EHCPool proposes the first 'Edge-to-node' score evaluation criterion based on edge features to assess node feature significance. To more effectively capture the critical subgraphs, a novel Iteration n-top strategy is further designed to adaptively learn sparse hard clustering assignments for graphs. Subsequently, an innovative N-E Aggregation strategy is presented to aggregate node and edge feature information in each independent subgraph. The proposed model was evaluated on multi-site brain imaging public datasets and yielded state-of-the-art performance. We believe this method is the first deep learning tool with the potential to probe different types of abnormal functional brain networks from data-driven perspective. Core code is at: https://github.com/swfen/EHCPool.
Unleashing the Strengths of Unlabeled Data in Pan-cancer Abdominal Organ Quantification: the FLARE22 Challenge
Aug 10, 2023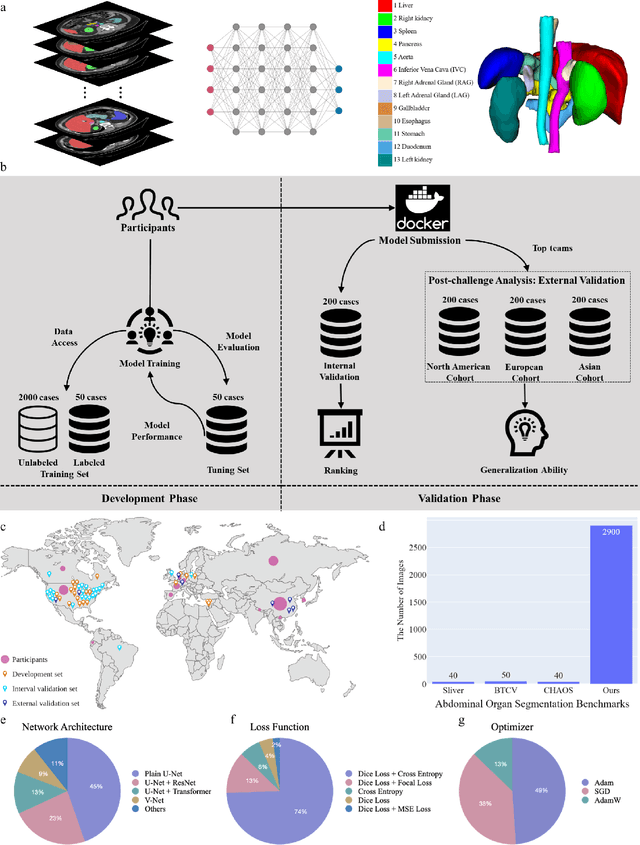
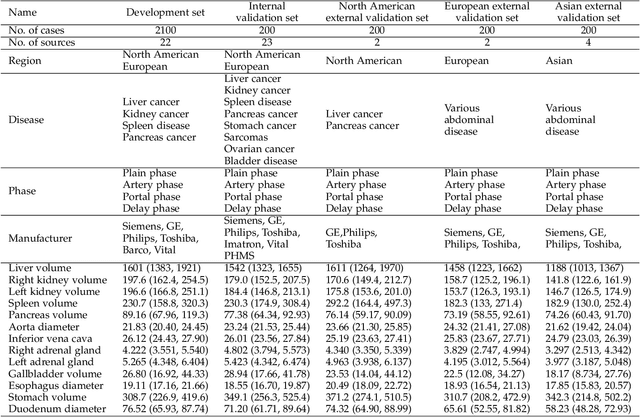
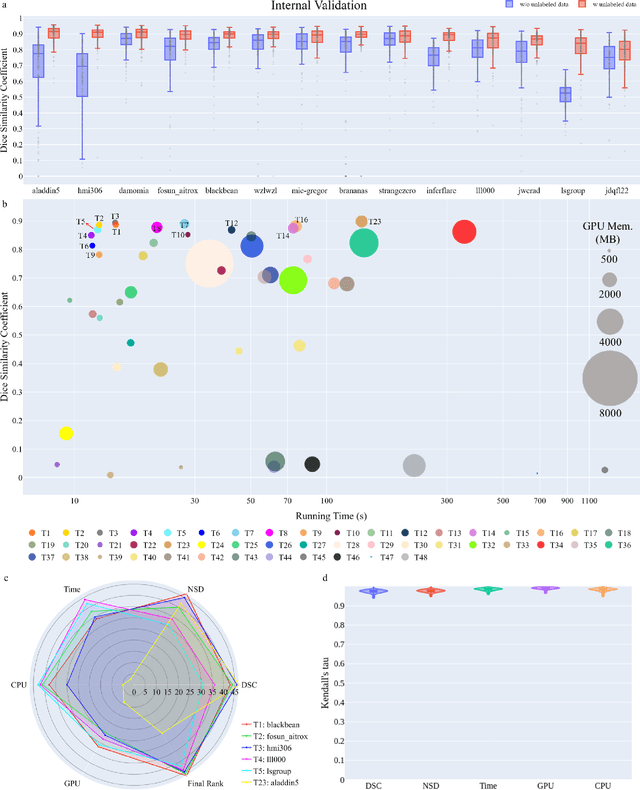
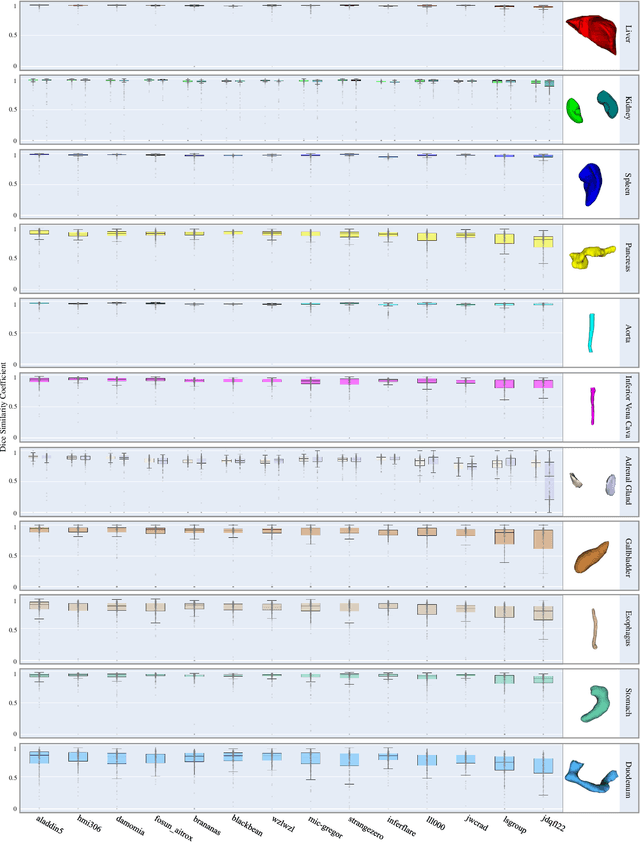
Abstract:Quantitative organ assessment is an essential step in automated abdominal disease diagnosis and treatment planning. Artificial intelligence (AI) has shown great potential to automatize this process. However, most existing AI algorithms rely on many expert annotations and lack a comprehensive evaluation of accuracy and efficiency in real-world multinational settings. To overcome these limitations, we organized the FLARE 2022 Challenge, the largest abdominal organ analysis challenge to date, to benchmark fast, low-resource, accurate, annotation-efficient, and generalized AI algorithms. We constructed an intercontinental and multinational dataset from more than 50 medical groups, including Computed Tomography (CT) scans with different races, diseases, phases, and manufacturers. We independently validated that a set of AI algorithms achieved a median Dice Similarity Coefficient (DSC) of 90.0\% by using 50 labeled scans and 2000 unlabeled scans, which can significantly reduce annotation requirements. The best-performing algorithms successfully generalized to holdout external validation sets, achieving a median DSC of 89.5\%, 90.9\%, and 88.3\% on North American, European, and Asian cohorts, respectively. They also enabled automatic extraction of key organ biology features, which was labor-intensive with traditional manual measurements. This opens the potential to use unlabeled data to boost performance and alleviate annotation shortages for modern AI models.
Controllable Multi-Objective Re-ranking with Policy Hypernetworks
Jun 13, 2023



Abstract:Multi-stage ranking pipelines have become widely used strategies in modern recommender systems, where the final stage aims to return a ranked list of items that balances a number of requirements such as user preference, diversity, novelty etc. Linear scalarization is arguably the most widely used technique to merge multiple requirements into one optimization objective, by summing up the requirements with certain preference weights. Existing final-stage ranking methods often adopt a static model where the preference weights are determined during offline training and kept unchanged during online serving. Whenever a modification of the preference weights is needed, the model has to be re-trained, which is time and resources inefficient. Meanwhile, the most appropriate weights may vary greatly for different groups of targeting users or at different time periods (e.g., during holiday promotions). In this paper, we propose a framework called controllable multi-objective re-ranking (CMR) which incorporates a hypernetwork to generate parameters for a re-ranking model according to different preference weights. In this way, CMR is enabled to adapt the preference weights according to the environment changes in an online manner, without retraining the models. Moreover, we classify practical business-oriented tasks into four main categories and seamlessly incorporate them in a new proposed re-ranking model based on an Actor-Evaluator framework, which serves as a reliable real-world testbed for CMR. Offline experiments based on the dataset collected from Taobao App showed that CMR improved several popular re-ranking models by using them as underlying models. Online A/B tests also demonstrated the effectiveness and trustworthiness of CMR.
Temporal Dynamic Synchronous Functional Brain Network for Schizophrenia Diagnosis and Lateralization Analysis
Apr 06, 2023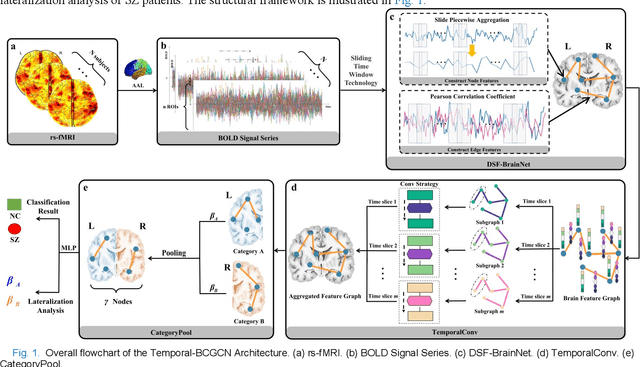
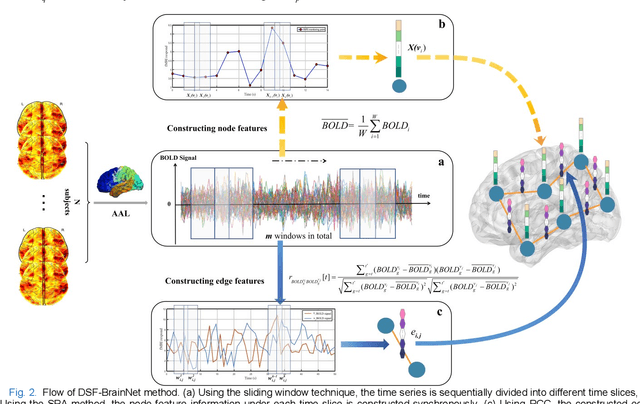
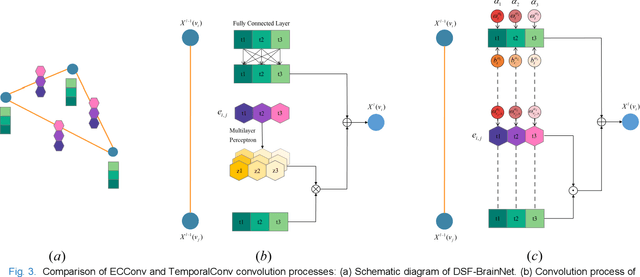
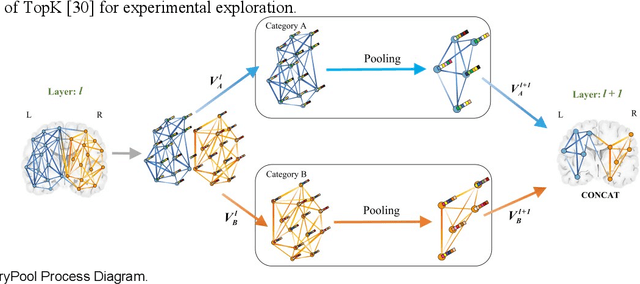
Abstract:The available evidence suggests that dynamic functional connectivity (dFC) can capture time-varying abnormalities in brain activity in resting-state cerebral functional magnetic resonance imaging (rs-fMRI) data and has a natural advantage in uncovering mechanisms of abnormal brain activity in schizophrenia(SZ) patients. Hence, an advanced dynamic brain network analysis model called the temporal brain category graph convolutional network (Temporal-BCGCN) was employed. Firstly, a unique dynamic brain network analysis module, DSF-BrainNet, was designed to construct dynamic synchronization features. Subsequently, a revolutionary graph convolution method, TemporalConv, was proposed, based on the synchronous temporal properties of feature. Finally, the first modular abnormal hemispherical lateralization test tool in deep learning based on rs-fMRI data, named CategoryPool, was proposed. This study was validated on COBRE and UCLA datasets and achieved 83.62% and 89.71% average accuracies, respectively, outperforming the baseline model and other state-of-the-art methods. The ablation results also demonstrate the advantages of TemporalConv over the traditional edge feature graph convolution approach and the improvement of CategoryPool over the classical graph pooling approach. Interestingly, this study showed that the lower order perceptual system and higher order network regions in the left hemisphere are more severely dysfunctional than in the right hemisphere in SZ and reaffirms the importance of the left medial superior frontal gyrus in SZ. Our core code is available at: https://github.com/swfen/Temporal-BCGCN.
AbdomenCT-1K: Is Abdominal Organ Segmentation A Solved Problem?
Oct 28, 2020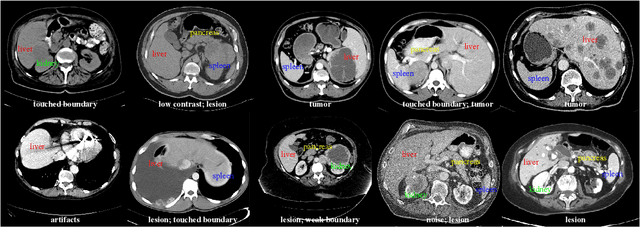
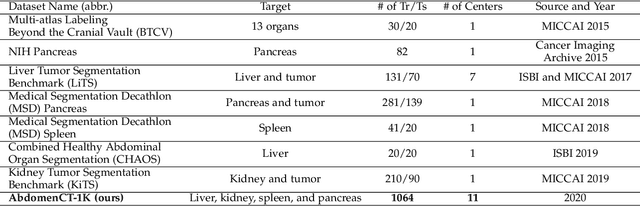
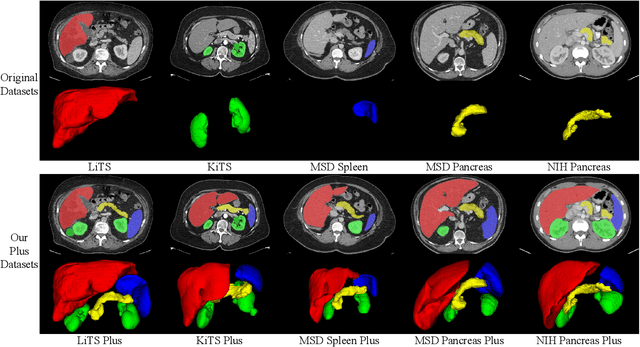
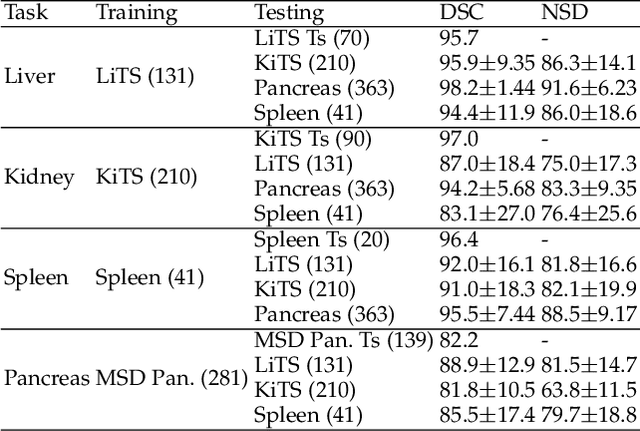
Abstract:With the unprecedented developments in deep learning, automatic segmentation of main abdominal organs (i.e., liver, kidney, and spleen) seems to be a solved problem as the state-of-the-art (SOTA) methods have achieved comparable results with inter-observer variability on existing benchmark datasets. However, most of the existing abdominal organ segmentation benchmark datasets only contain single-center, single-phase, single-vendor, or single-disease cases, thus, it is unclear whether the excellent performance can generalize on more diverse datasets. In this paper, we present a large and diverse abdominal CT organ segmentation dataset, termed as AbdomenCT-1K, with more than 1000 (1K) CT scans from 11 countries, including multi-center, multi-phase, multi-vendor, and multi-disease cases. Furthermore, we conduct a large-scale study for liver, kidney, spleen, and pancreas segmentation, as well as reveal the unsolved segmentation problems of the SOTA method, such as the limited generalization ability on distinct medical centers, phases, and unseen diseases. To advance the unsolved problems, we build four organ segmentation benchmarks for fully supervised, semi-supervised, weakly supervised, and continual learning, which are currently challenging and active research topics. Accordingly, we develop a simple and effective method for each benchmark, which can be used as out-of-the-box methods and strong baselines. We believe the introduction of the AbdomenCT-1K dataset will promote future in-depth research towards clinical applicable abdominal organ segmentation methods. Moreover, the datasets, codes, and trained models of baseline methods will be publicly available at https://github.com/JunMa11/AbdomenCT-1K.
Learning from Suspected Target: Bootstrapping Performance for Breast Cancer Detection in Mammography
Mar 01, 2020



Abstract:Deep learning object detection algorithm has been widely used in medical image analysis. Currently all the object detection tasks are based on the data annotated with object classes and their bounding boxes. On the other hand, medical images such as mammography usually contain normal regions or objects that are similar to the lesion region, and may be misclassified in the testing stage if they are not taken care of. In this paper, we address such problem by introducing a novel top likelihood loss together with a new sampling procedure to select and train the suspected target regions, as well as proposing a similarity loss to further identify suspected targets from targets. Mean average precision (mAP) according to the predicted targets and specificity, sensitivity, accuracy, AUC values according to classification of patients are adopted for performance comparisons. We firstly test our proposed method on a private dense mammogram dataset. Results show that our proposed method greatly reduce the false positive rate and the specificity is increased by 0.25 on detecting mass type cancer. It is worth mention that dense breast typically has a higher risk for developing breast cancers and also are harder for cancer detection in diagnosis, and our method outperforms a reported result from performance of radiologists. Our method is also validated on the public Digital Database for Screening Mammography (DDSM) dataset, brings significant improvement on mass type cancer detection and outperforms the most state-of-the-art work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge