Jian He
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
An Integrated Artificial Intelligence Operating System for Advanced Low-Altitude Aviation Applications
Nov 28, 2024Abstract:This paper introduces a comprehensive artificial intelligence operating system tailored for low-altitude aviation applications, integrating cutting-edge technologies for enhanced performance, safety, and efficiency. The system comprises six core components: OrinFlight OS, a high-performance operating system optimized for real-time task execution; UnitedVision, a versatile visual processing module supporting advanced image analysis; UnitedSense, a multi-sensor fusion module providing precise environmental modeling; UnitedNavigator, a dynamic path-planning and navigation system; UnitedMatrix, enabling multi-drone coordination and task execution; and UnitedInSight, a ground station for monitoring and management. Complemented by the UA DevKit low-code platform, the system facilitates user-friendly customization and application development. Leveraging NVIDIA Orin's computational power and advanced AI algorithms, this system addresses complex challenges in modern aviation, offering robust solutions for navigation, perception, and collaborative operations. This work highlights the system's architecture, features, and potential applications, demonstrating its ability to meet the demands of intelligent aviation environments.
Automatic Organ and Pan-cancer Segmentation in Abdomen CT: the FLARE 2023 Challenge
Aug 22, 2024Abstract:Organ and cancer segmentation in abdomen Computed Tomography (CT) scans is the prerequisite for precise cancer diagnosis and treatment. Most existing benchmarks and algorithms are tailored to specific cancer types, limiting their ability to provide comprehensive cancer analysis. This work presents the first international competition on abdominal organ and pan-cancer segmentation by providing a large-scale and diverse dataset, including 4650 CT scans with various cancer types from over 40 medical centers. The winning team established a new state-of-the-art with a deep learning-based cascaded framework, achieving average Dice Similarity Coefficient scores of 92.3% for organs and 64.9% for lesions on the hidden multi-national testing set. The dataset and code of top teams are publicly available, offering a benchmark platform to drive further innovations https://codalab.lisn.upsaclay.fr/competitions/12239.
YZS-model: A Predictive Model for Organic Drug Solubility Based on Graph Convolutional Networks and Transformer-Attention
Jun 27, 2024Abstract:The accurate prediction of drug molecule solubility is essential for determining their therapeutic effectiveness and safety, influencing the drug's ADME processes. Traditional solubility prediction techniques often fail to capture the complex nature of molecular tructures, leading to notable deviations between predictions and actual results. For example, the Discussion on Advanced Drug-Like Compound Structures. Lusci highlighted issues in capturing crucial cyclic structural information in molecules with ring structures. To overcome this issue, our research introduces a novel deep learning framework combining attention-based transformers, Long Short-Term Memory (LSTM) networks, and Graph Convolutional Networks (GCN), aimed at enhancing the precision of solubility predictions. Utilizing a training set of 9,943 compounds and testing on an anticancer compound dataset, our method achieved a correlation coefficient ($R^2$) of 0.55 and a Root Mean Square Error (RMSE) of 0.59, which outperforms the benchmark models' scores of 0.52 ($R^2$) and 0.61 (RMSE). Importantly, in an additional independent test, our model significantly outperformed the baseline with an RMSE of 1.05 compared to 1.28, a relative accuracy improvement of 45.9%. This research not only demonstrates the vast potential of deep learning for improving solubility prediction accuracy but also offers novel insights for drug design and selection in the future. Continued efforts will be directed towards optimizing the model architecture and extending its application to better support the drug development process, underscoring the pivotal role of deep learning in drug discovery.
Unleashing the Strengths of Unlabeled Data in Pan-cancer Abdominal Organ Quantification: the FLARE22 Challenge
Aug 10, 2023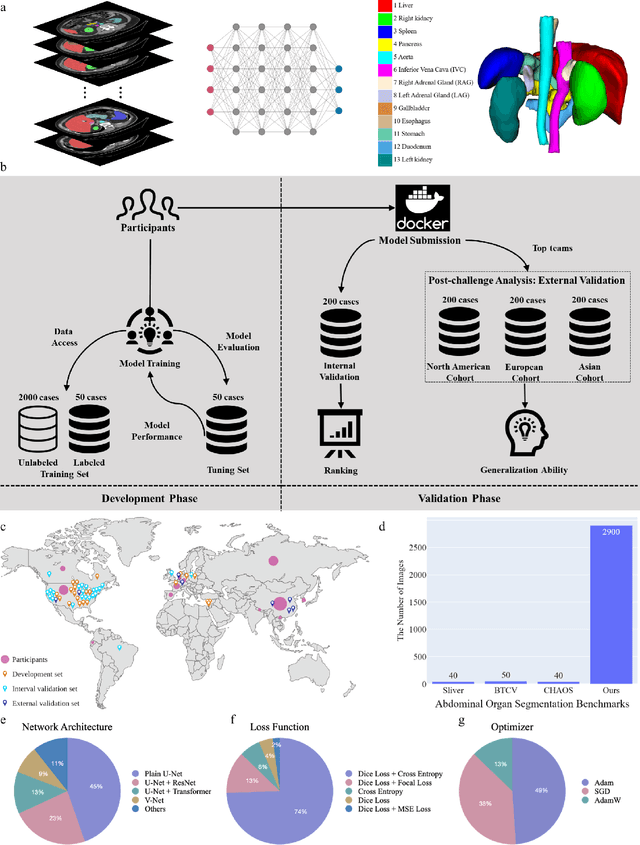
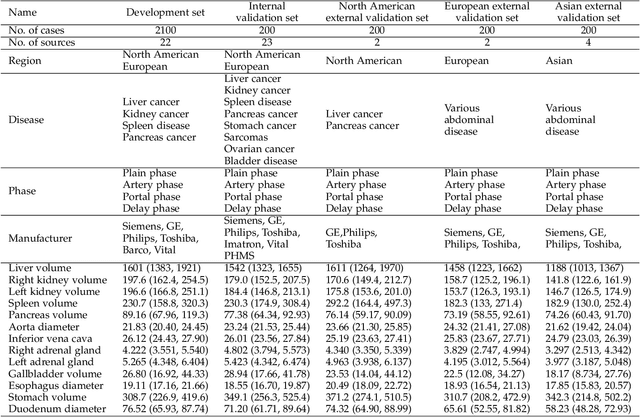
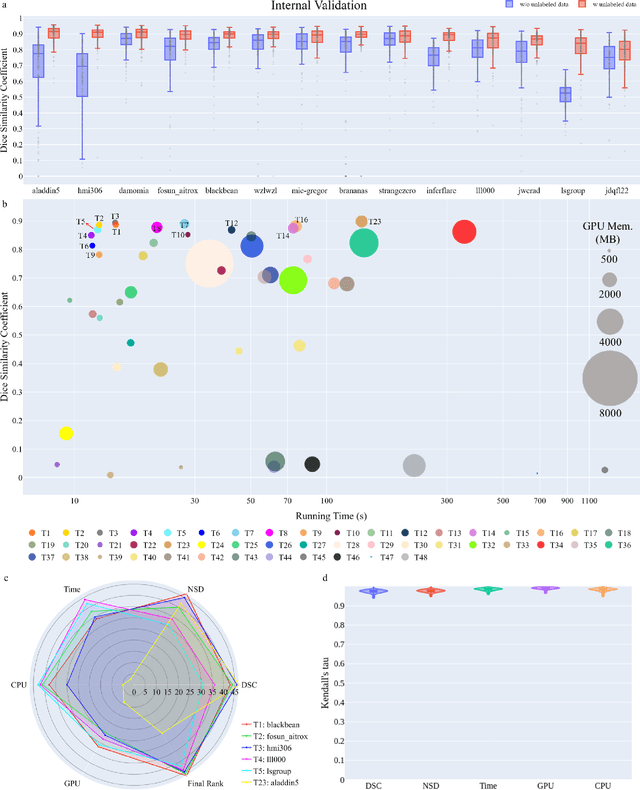
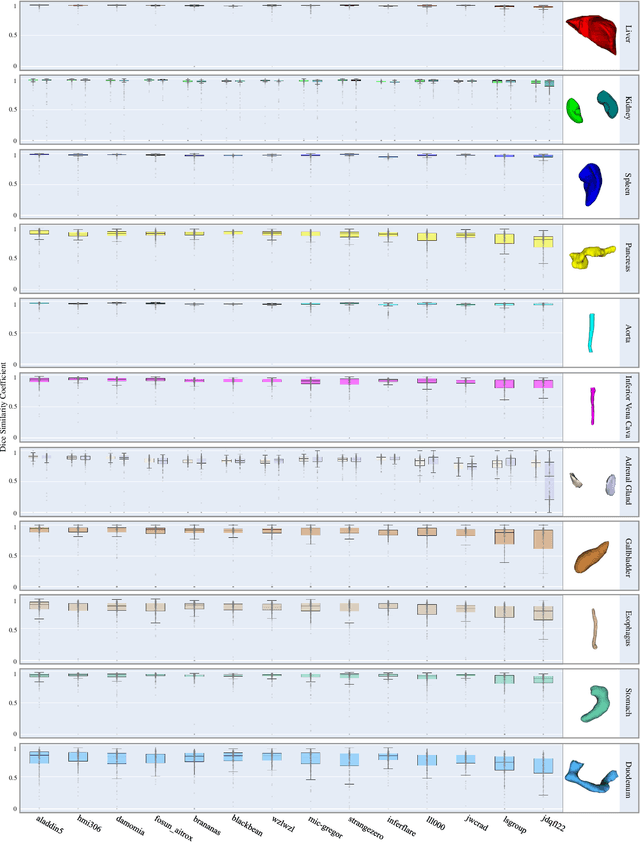
Abstract:Quantitative organ assessment is an essential step in automated abdominal disease diagnosis and treatment planning. Artificial intelligence (AI) has shown great potential to automatize this process. However, most existing AI algorithms rely on many expert annotations and lack a comprehensive evaluation of accuracy and efficiency in real-world multinational settings. To overcome these limitations, we organized the FLARE 2022 Challenge, the largest abdominal organ analysis challenge to date, to benchmark fast, low-resource, accurate, annotation-efficient, and generalized AI algorithms. We constructed an intercontinental and multinational dataset from more than 50 medical groups, including Computed Tomography (CT) scans with different races, diseases, phases, and manufacturers. We independently validated that a set of AI algorithms achieved a median Dice Similarity Coefficient (DSC) of 90.0\% by using 50 labeled scans and 2000 unlabeled scans, which can significantly reduce annotation requirements. The best-performing algorithms successfully generalized to holdout external validation sets, achieving a median DSC of 89.5\%, 90.9\%, and 88.3\% on North American, European, and Asian cohorts, respectively. They also enabled automatic extraction of key organ biology features, which was labor-intensive with traditional manual measurements. This opens the potential to use unlabeled data to boost performance and alleviate annotation shortages for modern AI models.
Adaptive Scheduling for Edge-Assisted DNN Serving
May 02, 2023Abstract:Deep neural networks (DNNs) have been widely used in various video analytic tasks. These tasks demand real-time responses. Due to the limited processing power on mobile devices, a common way to support such real-time analytics is to offload the processing to an edge server. This paper examines how to speed up the edge server DNN processing for multiple clients. In particular, we observe batching multiple DNN requests significantly speeds up the processing time. Based on this observation, we first design a novel scheduling algorithm to exploit the batching benefits of all requests that run the same DNN. This is compelling since there are only a handful of DNNs and many requests tend to use the same DNN. Our algorithms are general and can support different objectives, such as minimizing the completion time or maximizing the on-time ratio. We then extend our algorithm to handle requests that use different DNNs with or without shared layers. Finally, we develop a collaborative approach to further improve performance by adaptively processing some of the requests or portions of the requests locally at the clients. This is especially useful when the network and/or server is congested. Our implementation shows the effectiveness of our approach under different request distributions (e.g., Poisson, Pareto, and Constant inter-arrivals).
AbdomenCT-1K: Is Abdominal Organ Segmentation A Solved Problem?
Oct 28, 2020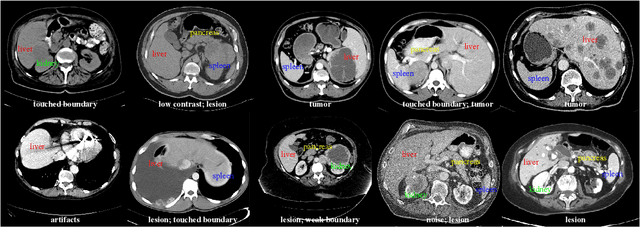
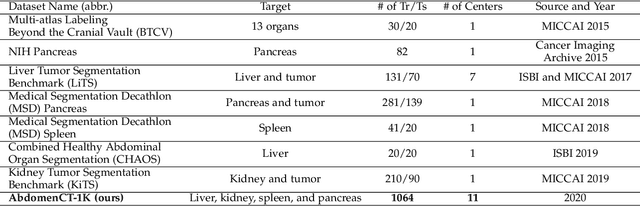
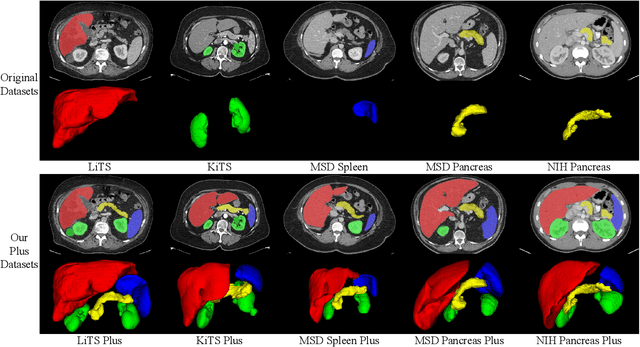
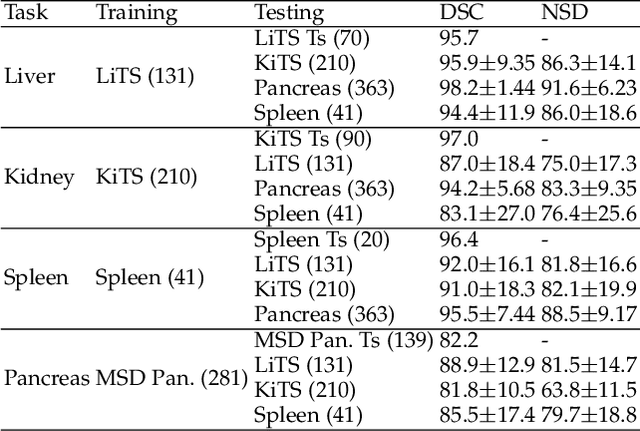
Abstract:With the unprecedented developments in deep learning, automatic segmentation of main abdominal organs (i.e., liver, kidney, and spleen) seems to be a solved problem as the state-of-the-art (SOTA) methods have achieved comparable results with inter-observer variability on existing benchmark datasets. However, most of the existing abdominal organ segmentation benchmark datasets only contain single-center, single-phase, single-vendor, or single-disease cases, thus, it is unclear whether the excellent performance can generalize on more diverse datasets. In this paper, we present a large and diverse abdominal CT organ segmentation dataset, termed as AbdomenCT-1K, with more than 1000 (1K) CT scans from 11 countries, including multi-center, multi-phase, multi-vendor, and multi-disease cases. Furthermore, we conduct a large-scale study for liver, kidney, spleen, and pancreas segmentation, as well as reveal the unsolved segmentation problems of the SOTA method, such as the limited generalization ability on distinct medical centers, phases, and unseen diseases. To advance the unsolved problems, we build four organ segmentation benchmarks for fully supervised, semi-supervised, weakly supervised, and continual learning, which are currently challenging and active research topics. Accordingly, we develop a simple and effective method for each benchmark, which can be used as out-of-the-box methods and strong baselines. We believe the introduction of the AbdomenCT-1K dataset will promote future in-depth research towards clinical applicable abdominal organ segmentation methods. Moreover, the datasets, codes, and trained models of baseline methods will be publicly available at https://github.com/JunMa11/AbdomenCT-1K.
Towards Efficient COVID-19 CT Annotation: A Benchmark for Lung and Infection Segmentation
Apr 27, 2020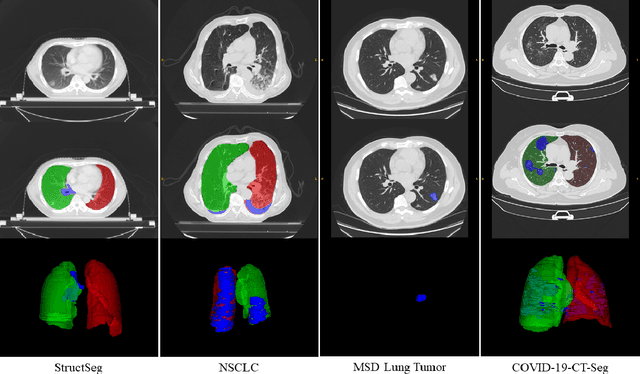

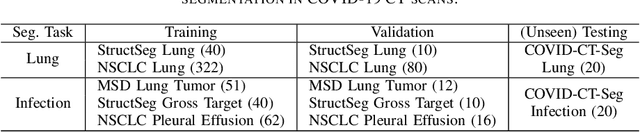
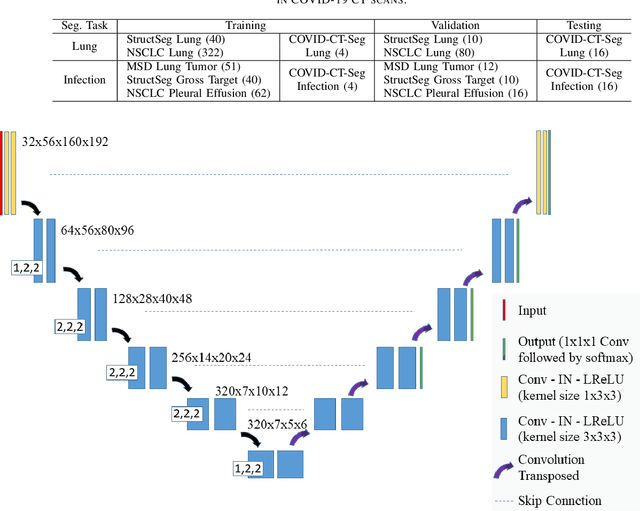
Abstract:Accurate segmentation of lung and infection in COVID-19 CT scans plays an important role in the quantitative management of patients. Most of the existing studies are based on large and private annotated datasets that are impractical to obtain from a single institution, especially when radiologists are busy fighting the coronavirus disease. Furthermore, it is hard to compare current COVID-19 CT segmentation methods as they are developed on different datasets, trained in different settings, and evaluated with different metrics. In this paper, we created a COVID-19 3D CT dataset with 20 cases that contains 1800+ annotated slices and made it publicly available. To promote the development of annotation-efficient deep learning methods, we built three benchmarks for lung and infection segmentation that contain current main research interests, e.g., few-shot learning, domain generalization, and knowledge transfer. For a fair comparison among different segmentation methods, we also provide unified training, validation and testing dataset splits, and evaluation metrics and corresponding code. In addition, we provided more than 40 pre-trained baseline models for the benchmarks, which not only serve as out-of-the-box segmentation tools but also save computational time for researchers who are interested in COVID-19 lung and infection segmentation. To the best of our knowledge, this work presents the largest public annotated COVID-19 CT volume dataset, the first segmentation benchmark, and the most pre-trained models up to now. We hope these resources (\url{https://gitee.com/junma11/COVID-19-CT-Seg-Benchmark}) could advance the development of deep learning methods for COVID-19 CT segmentation with limited data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge