Ziqi Yu
HiFi-Syn: Hierarchical Granularity Discrimination for High-Fidelity Synthesis of MR Images with Structure Preservation
Nov 21, 2023



Abstract:Synthesizing medical images while preserving their structural information is crucial in medical research. In such scenarios, the preservation of anatomical content becomes especially important. Although recent advances have been made by incorporating instance-level information to guide translation, these methods overlook the spatial coherence of structural-level representation and the anatomical invariance of content during translation. To address these issues, we introduce hierarchical granularity discrimination, which exploits various levels of semantic information present in medical images. Our strategy utilizes three levels of discrimination granularity: pixel-level discrimination using a Brain Memory Bank, structure-level discrimination on each brain structure with a re-weighting strategy to focus on hard samples, and global-level discrimination to ensure anatomical consistency during translation. The image translation performance of our strategy has been evaluated on three independent datasets (UK Biobank, IXI, and BraTS 2018), and it has outperformed state-of-the-art algorithms. Particularly, our model excels not only in synthesizing normal structures but also in handling abnormal (pathological) structures, such as brain tumors, despite the variations in contrast observed across different imaging modalities due to their pathological characteristics. The diagnostic value of synthesized MR images containing brain tumors has been evaluated by radiologists. This indicates that our model may offer an alternative solution in scenarios where specific MR modalities of patients are unavailable. Extensive experiments further demonstrate the versatility of our method, providing unique insights into medical image translation.
EFX Allocations Exist for Binary Valuations
Aug 10, 2023Abstract:We study the fair division problem and the existence of allocations satisfying the fairness criterion envy-freeness up to any item (EFX). The existence of EFX allocations is a major open problem in the fair division literature. We consider binary valuations where the marginal gain of the value by receiving an extra item is either $0$ or $1$. Babaioff et al. [2021] proved that EFX allocations always exist for binary and submodular valuations. In this paper, by using completely different techniques, we extend this existence result to general binary valuations that are not necessarily submodular, and we present a polynomial time algorithm for computing an EFX allocation.
MouseGAN++: Unsupervised Disentanglement and Contrastive Representation for Multiple MRI Modalities Synthesis and Structural Segmentation of Mouse Brain
Dec 04, 2022



Abstract:Segmenting the fine structure of the mouse brain on magnetic resonance (MR) images is critical for delineating morphological regions, analyzing brain function, and understanding their relationships. Compared to a single MRI modality, multimodal MRI data provide complementary tissue features that can be exploited by deep learning models, resulting in better segmentation results. However, multimodal mouse brain MRI data is often lacking, making automatic segmentation of mouse brain fine structure a very challenging task. To address this issue, it is necessary to fuse multimodal MRI data to produce distinguished contrasts in different brain structures. Hence, we propose a novel disentangled and contrastive GAN-based framework, named MouseGAN++, to synthesize multiple MR modalities from single ones in a structure-preserving manner, thus improving the segmentation performance by imputing missing modalities and multi-modality fusion. Our results demonstrate that the translation performance of our method outperforms the state-of-the-art methods. Using the subsequently learned modality-invariant information as well as the modality-translated images, MouseGAN++ can segment fine brain structures with averaged dice coefficients of 90.0% (T2w) and 87.9% (T1w), respectively, achieving around +10% performance improvement compared to the state-of-the-art algorithms. Our results demonstrate that MouseGAN++, as a simultaneous image synthesis and segmentation method, can be used to fuse cross-modality information in an unpaired manner and yield more robust performance in the absence of multimodal data. We release our method as a mouse brain structural segmentation tool for free academic usage at https://github.com/yu02019.
Towards Efficient COVID-19 CT Annotation: A Benchmark for Lung and Infection Segmentation
Apr 27, 2020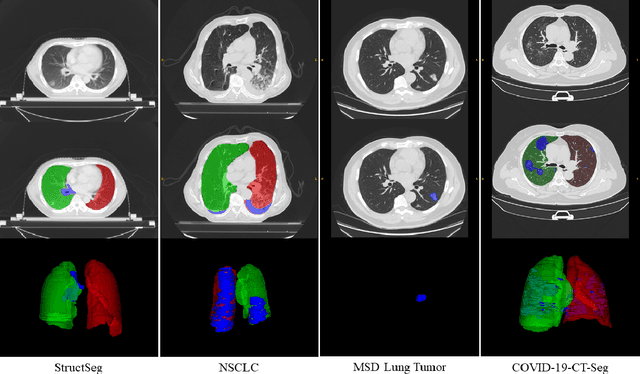

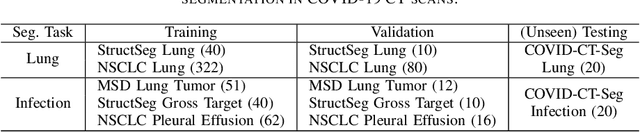
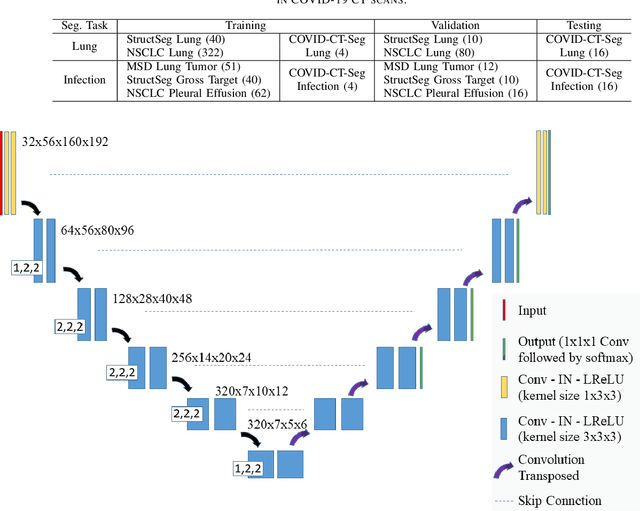
Abstract:Accurate segmentation of lung and infection in COVID-19 CT scans plays an important role in the quantitative management of patients. Most of the existing studies are based on large and private annotated datasets that are impractical to obtain from a single institution, especially when radiologists are busy fighting the coronavirus disease. Furthermore, it is hard to compare current COVID-19 CT segmentation methods as they are developed on different datasets, trained in different settings, and evaluated with different metrics. In this paper, we created a COVID-19 3D CT dataset with 20 cases that contains 1800+ annotated slices and made it publicly available. To promote the development of annotation-efficient deep learning methods, we built three benchmarks for lung and infection segmentation that contain current main research interests, e.g., few-shot learning, domain generalization, and knowledge transfer. For a fair comparison among different segmentation methods, we also provide unified training, validation and testing dataset splits, and evaluation metrics and corresponding code. In addition, we provided more than 40 pre-trained baseline models for the benchmarks, which not only serve as out-of-the-box segmentation tools but also save computational time for researchers who are interested in COVID-19 lung and infection segmentation. To the best of our knowledge, this work presents the largest public annotated COVID-19 CT volume dataset, the first segmentation benchmark, and the most pre-trained models up to now. We hope these resources (\url{https://gitee.com/junma11/COVID-19-CT-Seg-Benchmark}) could advance the development of deep learning methods for COVID-19 CT segmentation with limited data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge