Henkjan Huisman
Optimizing Federated Learning Configurations for MRI Prostate Segmentation and Cancer Detection: A Simulation Study
Jul 30, 2025Abstract:Purpose: To develop and optimize a federated learning (FL) framework across multiple clients for biparametric MRI prostate segmentation and clinically significant prostate cancer (csPCa) detection. Materials and Methods: A retrospective study was conducted using Flower FL to train a nnU-Net-based architecture for MRI prostate segmentation and csPCa detection, using data collected from January 2010 to August 2021. Model development included training and optimizing local epochs, federated rounds, and aggregation strategies for FL-based prostate segmentation on T2-weighted MRIs (four clients, 1294 patients) and csPCa detection using biparametric MRIs (three clients, 1440 patients). Performance was evaluated on independent test sets using the Dice score for segmentation and the Prostate Imaging: Cancer Artificial Intelligence (PI-CAI) score, defined as the average of the area under the receiver operating characteristic curve and average precision, for csPCa detection. P-values for performance differences were calculated using permutation testing. Results: The FL configurations were independently optimized for both tasks, showing improved performance at 1 epoch 300 rounds using FedMedian for prostate segmentation and 5 epochs 200 rounds using FedAdagrad, for csPCa detection. Compared with the average performance of the clients, the optimized FL model significantly improved performance in prostate segmentation and csPCa detection on the independent test set. The optimized FL model showed higher lesion detection performance compared to the FL-baseline model, but no evidence of a difference was observed for prostate segmentation. Conclusions: FL enhanced the performance and generalizability of MRI prostate segmentation and csPCa detection compared with local models, and optimizing its configuration further improved lesion detection performance.
Latent Diffusion Autoencoders: Toward Efficient and Meaningful Unsupervised Representation Learning in Medical Imaging
Apr 11, 2025Abstract:This study presents Latent Diffusion Autoencoder (LDAE), a novel encoder-decoder diffusion-based framework for efficient and meaningful unsupervised learning in medical imaging, focusing on Alzheimer disease (AD) using brain MR from the ADNI database as a case study. Unlike conventional diffusion autoencoders operating in image space, LDAE applies the diffusion process in a compressed latent representation, improving computational efficiency and making 3D medical imaging representation learning tractable. To validate the proposed approach, we explore two key hypotheses: (i) LDAE effectively captures meaningful semantic representations on 3D brain MR associated with AD and ageing, and (ii) LDAE achieves high-quality image generation and reconstruction while being computationally efficient. Experimental results support both hypotheses: (i) linear-probe evaluations demonstrate promising diagnostic performance for AD (ROC-AUC: 90%, ACC: 84%) and age prediction (MAE: 4.1 years, RMSE: 5.2 years); (ii) the learned semantic representations enable attribute manipulation, yielding anatomically plausible modifications; (iii) semantic interpolation experiments show strong reconstruction of missing scans, with SSIM of 0.969 (MSE: 0.0019) for a 6-month gap. Even for longer gaps (24 months), the model maintains robust performance (SSIM > 0.93, MSE < 0.004), indicating an ability to capture temporal progression trends; (iv) compared to conventional diffusion autoencoders, LDAE significantly increases inference throughput (20x faster) while also enhancing reconstruction quality. These findings position LDAE as a promising framework for scalable medical imaging applications, with the potential to serve as a foundation model for medical image analysis. Code available at https://github.com/GabrieleLozupone/LDAE
Deep Learning-based Unsupervised Domain Adaptation via a Unified Model for Prostate Lesion Detection Using Multisite Bi-parametric MRI Datasets
Aug 08, 2024


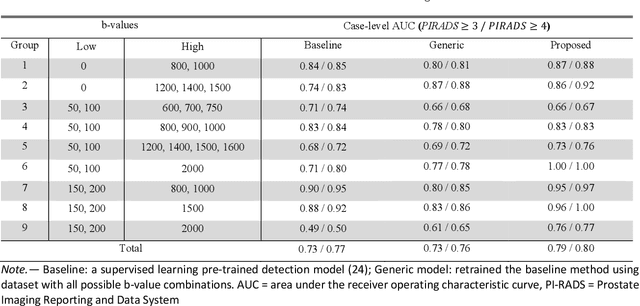
Abstract:Our hypothesis is that UDA using diffusion-weighted images, generated with a unified model, offers a promising and reliable strategy for enhancing the performance of supervised learning models in multi-site prostate lesion detection, especially when various b-values are present. This retrospective study included data from 5,150 patients (14,191 samples) collected across nine different imaging centers. A novel UDA method using a unified generative model was developed for multi-site PCa detection. This method translates diffusion-weighted imaging (DWI) acquisitions, including apparent diffusion coefficient (ADC) and individual DW images acquired using various b-values, to align with the style of images acquired using b-values recommended by Prostate Imaging Reporting and Data System (PI-RADS) guidelines. The generated ADC and DW images replace the original images for PCa detection. An independent set of 1,692 test cases (2,393 samples) was used for evaluation. The area under the receiver operating characteristic curve (AUC) was used as the primary metric, and statistical analysis was performed via bootstrapping. For all test cases, the AUC values for baseline SL and UDA methods were 0.73 and 0.79 (p<.001), respectively, for PI-RADS>=3, and 0.77 and 0.80 (p<.001) for PI-RADS>=4 PCa lesions. In the 361 test cases under the most unfavorable image acquisition setting, the AUC values for baseline SL and UDA were 0.49 and 0.76 (p<.001) for PI-RADS>=3, and 0.50 and 0.77 (p<.001) for PI-RADS>=4 PCa lesions. The results indicate the proposed UDA with generated images improved the performance of SL methods in multi-site PCa lesion detection across datasets with various b values, especially for images acquired with significant deviations from the PI-RADS recommended DWI protocol (e.g. with an extremely high b-value).
Deformable MRI Sequence Registration for AI-based Prostate Cancer Diagnosis
Apr 15, 2024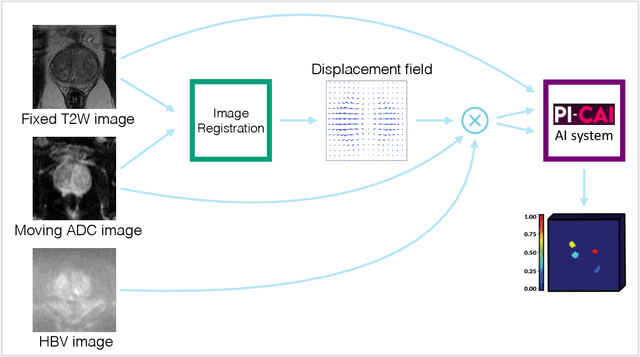
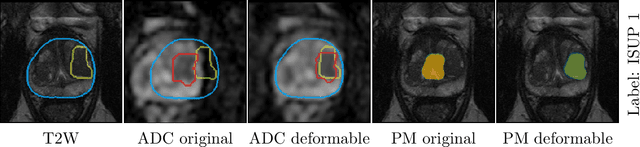
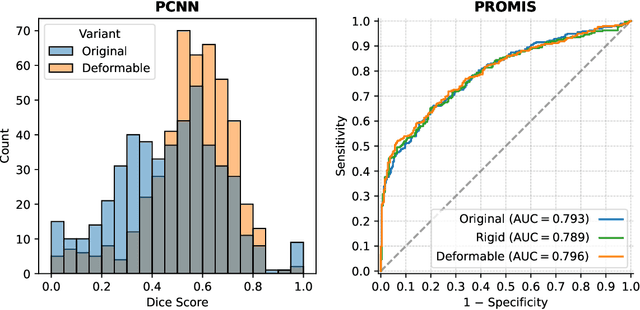
Abstract:The PI-CAI (Prostate Imaging: Cancer AI) challenge led to expert-level diagnostic algorithms for clinically significant prostate cancer detection. The algorithms receive biparametric MRI scans as input, which consist of T2-weighted and diffusion-weighted scans. These scans can be misaligned due to multiple factors in the scanning process. Image registration can alleviate this issue by predicting the deformation between the sequences. We investigate the effect of image registration on the diagnostic performance of AI-based prostate cancer diagnosis. First, the image registration algorithm, developed in MeVisLab, is analyzed using a dataset with paired lesion annotations. Second, the effect on diagnosis is evaluated by comparing case-level cancer diagnosis performance between using the original dataset, rigidly aligned diffusion-weighted scans, or deformably aligned diffusion-weighted scans. Rigid registration showed no improvement. Deformable registration demonstrated a substantial improvement in lesion overlap (+10% median Dice score) and a positive yet non-significant improvement in diagnostic performance (+0.3% AUROC, p=0.18). Our investigation shows that a substantial improvement in lesion alignment does not directly lead to a significant improvement in diagnostic performance. Qualitative analysis indicated that jointly developing image registration methods and diagnostic AI algorithms could enhance diagnostic accuracy and patient outcomes.
Uncertainty-Aware Semi-Supervised Learning for Prostate MRI Zonal Segmentation
May 10, 2023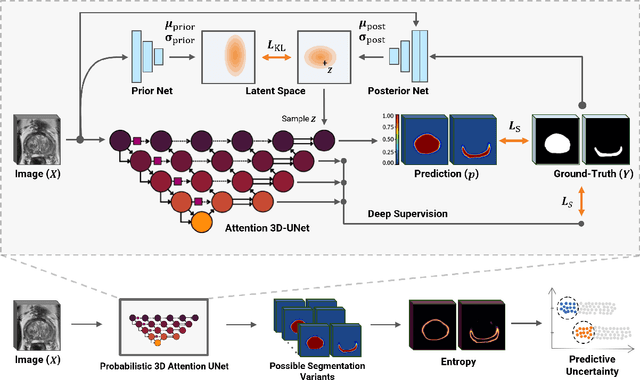
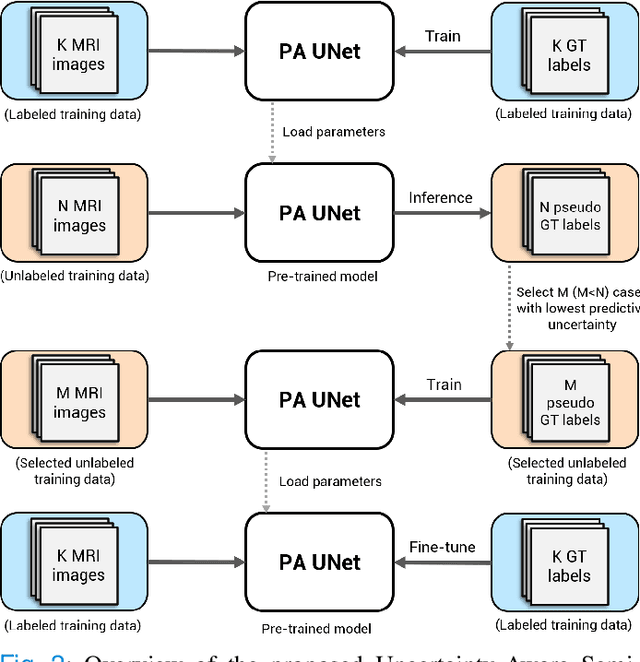
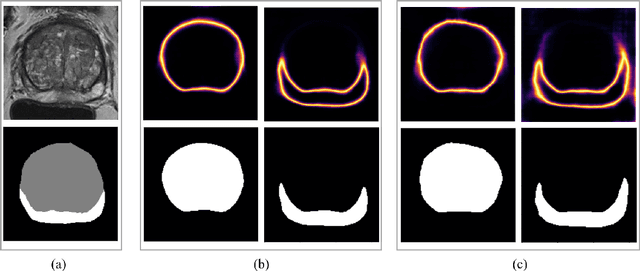
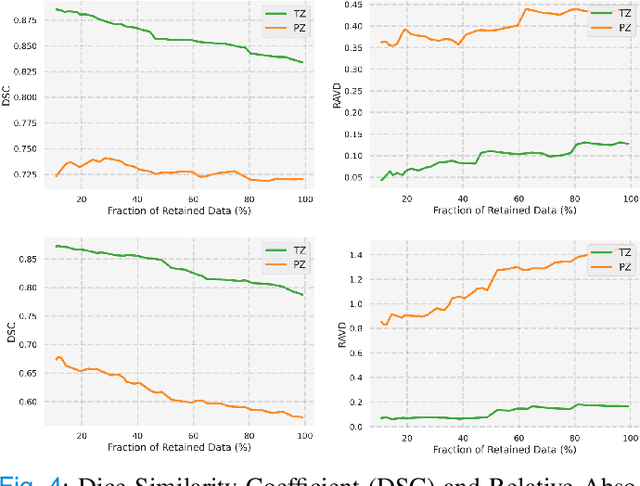
Abstract:Quality of deep convolutional neural network predictions strongly depends on the size of the training dataset and the quality of the annotations. Creating annotations, especially for 3D medical image segmentation, is time-consuming and requires expert knowledge. We propose a novel semi-supervised learning (SSL) approach that requires only a relatively small number of annotations while being able to use the remaining unlabeled data to improve model performance. Our method uses a pseudo-labeling technique that employs recent deep learning uncertainty estimation models. By using the estimated uncertainty, we were able to rank pseudo-labels and automatically select the best pseudo-annotations generated by the supervised model. We applied this to prostate zonal segmentation in T2-weighted MRI scans. Our proposed model outperformed the semi-supervised model in experiments with the ProstateX dataset and an external test set, by leveraging only a subset of unlabeled data rather than the full collection of 4953 cases, our proposed model demonstrated improved performance. The segmentation dice similarity coefficient in the transition zone and peripheral zone increased from 0.835 and 0.727 to 0.852 and 0.751, respectively, for fully supervised model and the uncertainty-aware semi-supervised learning model (USSL). Our USSL model demonstrates the potential to allow deep learning models to be trained on large datasets without requiring full annotation. Our code is available at https://github.com/DIAGNijmegen/prostateMR-USSL.
Medical diffusion on a budget: textual inversion for medical image generation
Mar 23, 2023



Abstract:Diffusion-based models for text-to-image generation have gained immense popularity due to recent advancements in efficiency, accessibility, and quality. Although it is becoming increasingly feasible to perform inference with these systems using consumer-grade GPUs, training them from scratch still requires access to large datasets and significant computational resources. In the case of medical image generation, the availability of large, publicly accessible datasets that include text reports is limited due to legal and ethical concerns. While training a diffusion model on a private dataset may address this issue, it is not always feasible for institutions lacking the necessary computational resources. This work demonstrates that pre-trained Stable Diffusion models, originally trained on natural images, can be adapted to various medical imaging modalities by training text embeddings with textual inversion. In this study, we conducted experiments using medical datasets comprising only 100 samples from three medical modalities. Embeddings were trained in a matter of hours, while still retaining diagnostic relevance in image generation. Experiments were designed to achieve several objectives. Firstly, we fine-tuned the training and inference processes of textual inversion, revealing that larger embeddings and more examples are required. Secondly, we validated our approach by demonstrating a 2\% increase in the diagnostic accuracy (AUC) for detecting prostate cancer on MRI, which is a challenging multi-modal imaging modality, from 0.78 to 0.80. Thirdly, we performed simulations by interpolating between healthy and diseased states, combining multiple pathologies, and inpainting to show embedding flexibility and control of disease appearance. Finally, the embeddings trained in this study are small (less than 1 MB), which facilitates easy sharing of medical data with reduced privacy concerns.
Prototypical few-shot segmentation for cross-institution male pelvic structures with spatial registration
Sep 13, 2022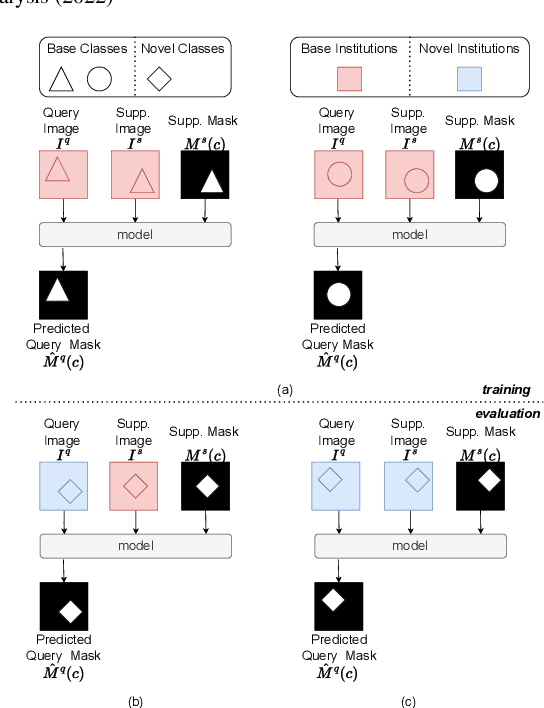

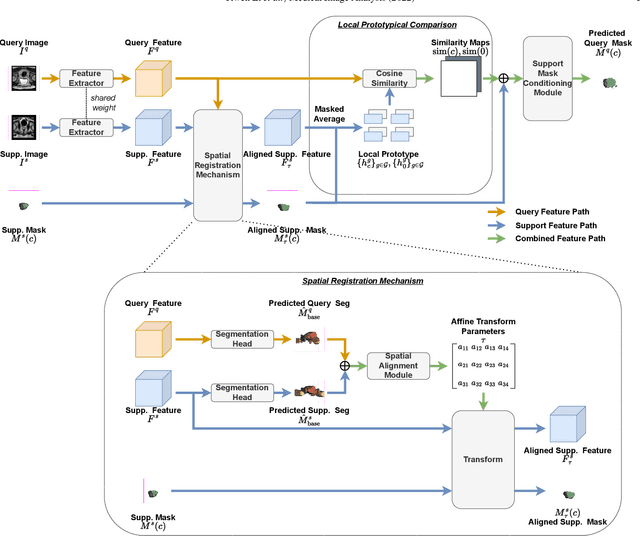
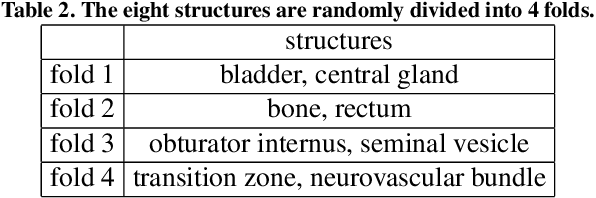
Abstract:The prowess that makes few-shot learning desirable in medical image analysis is the efficient use of the support image data, which are labelled to classify or segment new classes, a task that otherwise requires substantially more training images and expert annotations. This work describes a fully 3D prototypical few-shot segmentation algorithm, such that the trained networks can be effectively adapted to clinically interesting structures that are absent in training, using only a few labelled images from a different institute. First, to compensate for the widely recognised spatial variability between institutions in episodic adaptation of novel classes, a novel spatial registration mechanism is integrated into prototypical learning, consisting of a segmentation head and an spatial alignment module. Second, to assist the training with observed imperfect alignment, support mask conditioning module is proposed to further utilise the annotation available from the support images. Extensive experiments are presented in an application of segmenting eight anatomical structures important for interventional planning, using a data set of 589 pelvic T2-weighted MR images, acquired at seven institutes. The results demonstrate the efficacy in each of the 3D formulation, the spatial registration, and the support mask conditioning, all of which made positive contributions independently or collectively. Compared with the previously proposed 2D alternatives, the few-shot segmentation performance was improved with statistical significance, regardless whether the support data come from the same or different institutes.
Few-shot image segmentation for cross-institution male pelvic organs using registration-assisted prototypical learning
Jan 17, 2022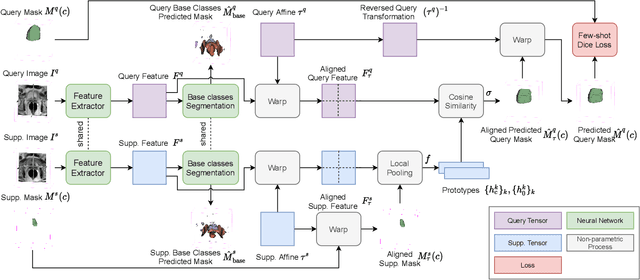
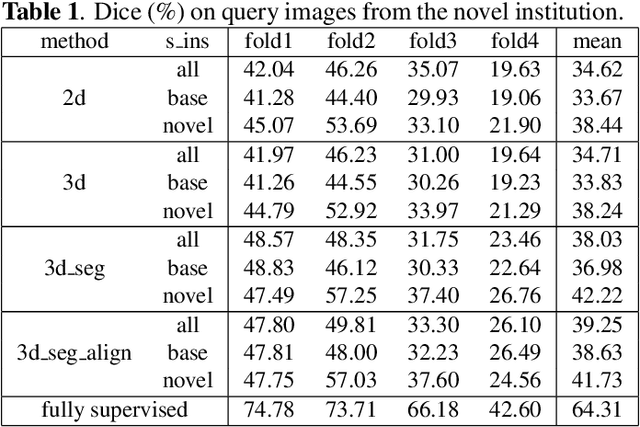
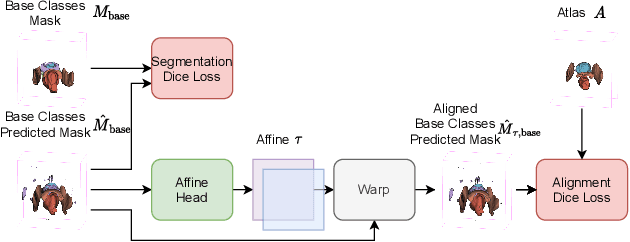
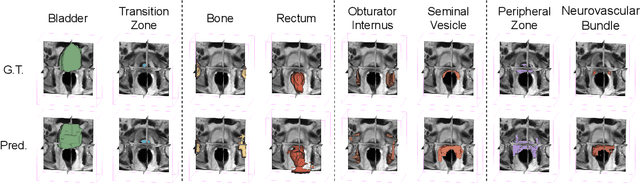
Abstract:The ability to adapt medical image segmentation networks for a novel class such as an unseen anatomical or pathological structure, when only a few labelled examples of this class are available from local healthcare providers, is sought-after. This potentially addresses two widely recognised limitations in deploying modern deep learning models to clinical practice, expertise-and-labour-intensive labelling and cross-institution generalisation. This work presents the first 3D few-shot interclass segmentation network for medical images, using a labelled multi-institution dataset from prostate cancer patients with eight regions of interest. We propose an image alignment module registering the predicted segmentation of both query and support data, in a standard prototypical learning algorithm, to a reference atlas space. The built-in registration mechanism can effectively utilise the prior knowledge of consistent anatomy between subjects, regardless whether they are from the same institution or not. Experimental results demonstrated that the proposed registration-assisted prototypical learning significantly improved segmentation accuracy (p-values<0.01) on query data from a holdout institution, with varying availability of support data from multiple institutions. We also report the additional benefits of the proposed 3D networks with 75% fewer parameters and an arguably simpler implementation, compared with existing 2D few-shot approaches that segment 2D slices of volumetric medical images.
Report-Guided Automatic Lesion Annotation for Deep Learning-Based Prostate Cancer Detection in bpMRI
Dec 09, 2021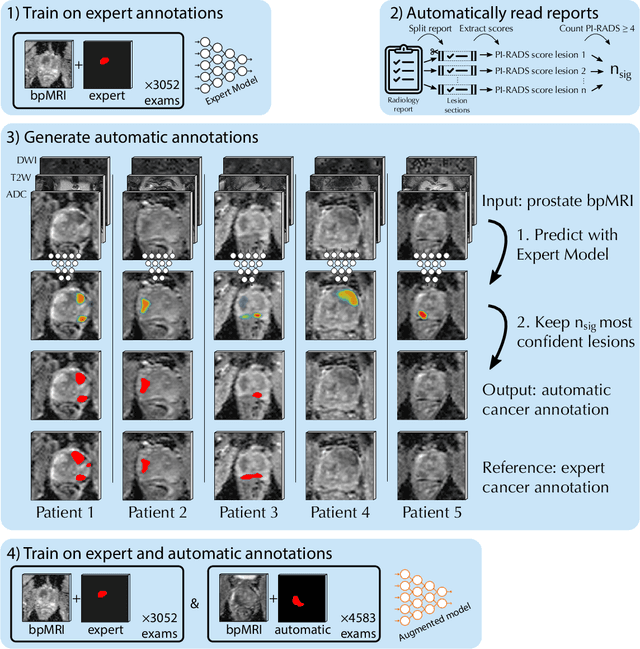

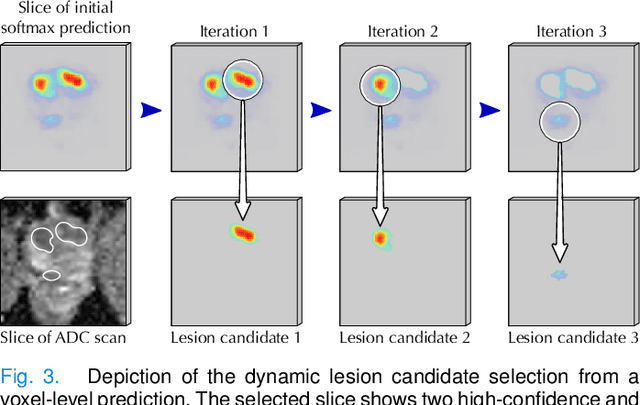
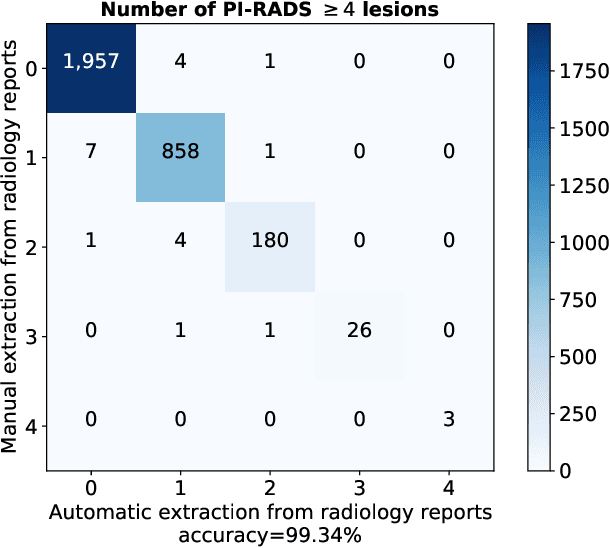
Abstract:Deep learning-based diagnostic performance increases with more annotated data, but manual annotation is a bottleneck in most fields. Experts evaluate diagnostic images during clinical routine, and write their findings in reports. Automatic annotation based on clinical reports could overcome the manual labelling bottleneck. We hypothesise that dense annotations for detection tasks can be generated using model predictions, guided by sparse information from these reports. To demonstrate efficacy, we generated clinically significant prostate cancer (csPCa) annotations, guided by the number of clinically significant findings in the radiology reports. We included 7,756 prostate MRI examinations, of which 3,050 were manually annotated and 4,706 were automatically annotated. We evaluated the automatic annotation quality on the manually annotated subset: our score extraction correctly identified the number of csPCa lesions for $99.3\%$ of the reports and our csPCa segmentation model correctly localised $83.8 \pm 1.1\%$ of the lesions. We evaluated prostate cancer detection performance on 300 exams from an external centre with histopathology-confirmed ground truth. Augmenting the training set with automatically labelled exams improved patient-based diagnostic area under the receiver operating characteristic curve from $88.1\pm 1.1\%$ to $89.8\pm 1.0\%$ ($P = 1.2 \cdot 10^{-4}$) and improved lesion-based sensitivity at one false positive per case from $79.2 \pm 2.8\%$ to $85.4 \pm 1.9\%$ ($P<10^{-4}$), with $mean \pm std.$ over 15 independent runs. This improved performance demonstrates the feasibility of our report-guided automatic annotations. Source code is made publicly available at https://github.com/DIAGNijmegen/Report-Guided-Annotation. Best csPCa detection algorithm is made available at https://grand-challenge.org/algorithms/bpmri-cspca-detection-report-guided-annotations/.
Fully Automatic Deep Learning Framework for Pancreatic Ductal Adenocarcinoma Detection on Computed Tomography
Dec 02, 2021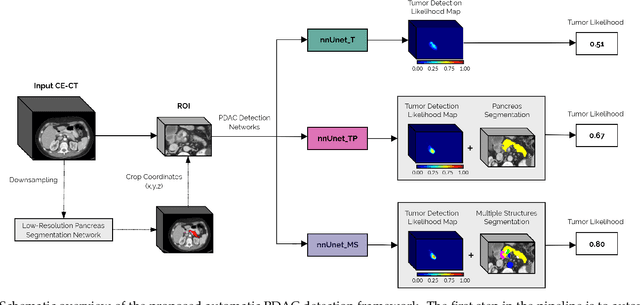

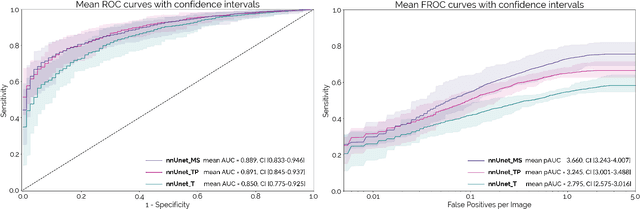
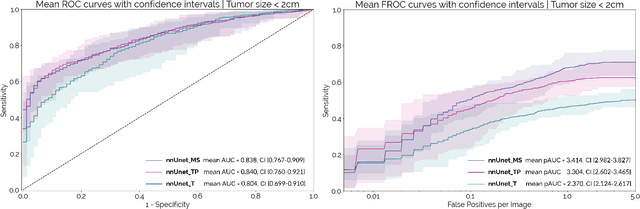
Abstract:Early detection improves prognosis in pancreatic ductal adenocarcinoma (PDAC) but is challenging as lesions are often small and poorly defined on contrast-enhanced computed tomography scans (CE-CT). Deep learning can facilitate PDAC diagnosis, however current models still fail to identify small (<2cm) lesions. In this study, state-of-the-art deep learning models were used to develop an automatic framework for PDAC detection, focusing on small lesions. Additionally, the impact of integrating surrounding anatomy was investigated. CE-CT scans from a cohort of 119 pathology-proven PDAC patients and a cohort of 123 patients without PDAC were used to train a nnUnet for automatic lesion detection and segmentation (nnUnet_T). Two additional nnUnets were trained to investigate the impact of anatomy integration: (1) segmenting the pancreas and tumor (nnUnet_TP), (2) segmenting the pancreas, tumor, and multiple surrounding anatomical structures (nnUnet_MS). An external, publicly available test set was used to compare the performance of the three networks. The nnUnet_MS achieved the best performance, with an area under the receiver operating characteristic curve of 0.91 for the whole test set and 0.88 for tumors <2cm, showing that state-of-the-art deep learning can detect small PDAC and benefits from anatomy information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge