Joeran S. Bosma
MHub.ai: A Simple, Standardized, and Reproducible Platform for AI Models in Medical Imaging
Jan 15, 2026Abstract:Artificial intelligence (AI) has the potential to transform medical imaging by automating image analysis and accelerating clinical research. However, research and clinical use are limited by the wide variety of AI implementations and architectures, inconsistent documentation, and reproducibility issues. Here, we introduce MHub.ai, an open-source, container-based platform that standardizes access to AI models with minimal configuration, promoting accessibility and reproducibility in medical imaging. MHub.ai packages models from peer-reviewed publications into standardized containers that support direct processing of DICOM and other formats, provide a unified application interface, and embed structured metadata. Each model is accompanied by publicly available reference data that can be used to confirm model operation. MHub.ai includes an initial set of state-of-the-art segmentation, prediction, and feature extraction models for different modalities. The modular framework enables adaptation of any model and supports community contributions. We demonstrate the utility of the platform in a clinical use case through comparative evaluation of lung segmentation models. To further strengthen transparency and reproducibility, we publicly release the generated segmentations and evaluation metrics and provide interactive dashboards that allow readers to inspect individual cases and reproduce or extend our analysis. By simplifying model use, MHub.ai enables side-by-side benchmarking with identical execution commands and standardized outputs, and lowers the barrier to clinical translation.
Optimizing Federated Learning Configurations for MRI Prostate Segmentation and Cancer Detection: A Simulation Study
Jul 30, 2025Abstract:Purpose: To develop and optimize a federated learning (FL) framework across multiple clients for biparametric MRI prostate segmentation and clinically significant prostate cancer (csPCa) detection. Materials and Methods: A retrospective study was conducted using Flower FL to train a nnU-Net-based architecture for MRI prostate segmentation and csPCa detection, using data collected from January 2010 to August 2021. Model development included training and optimizing local epochs, federated rounds, and aggregation strategies for FL-based prostate segmentation on T2-weighted MRIs (four clients, 1294 patients) and csPCa detection using biparametric MRIs (three clients, 1440 patients). Performance was evaluated on independent test sets using the Dice score for segmentation and the Prostate Imaging: Cancer Artificial Intelligence (PI-CAI) score, defined as the average of the area under the receiver operating characteristic curve and average precision, for csPCa detection. P-values for performance differences were calculated using permutation testing. Results: The FL configurations were independently optimized for both tasks, showing improved performance at 1 epoch 300 rounds using FedMedian for prostate segmentation and 5 epochs 200 rounds using FedAdagrad, for csPCa detection. Compared with the average performance of the clients, the optimized FL model significantly improved performance in prostate segmentation and csPCa detection on the independent test set. The optimized FL model showed higher lesion detection performance compared to the FL-baseline model, but no evidence of a difference was observed for prostate segmentation. Conclusions: FL enhanced the performance and generalizability of MRI prostate segmentation and csPCa detection compared with local models, and optimizing its configuration further improved lesion detection performance.
Deformable MRI Sequence Registration for AI-based Prostate Cancer Diagnosis
Apr 15, 2024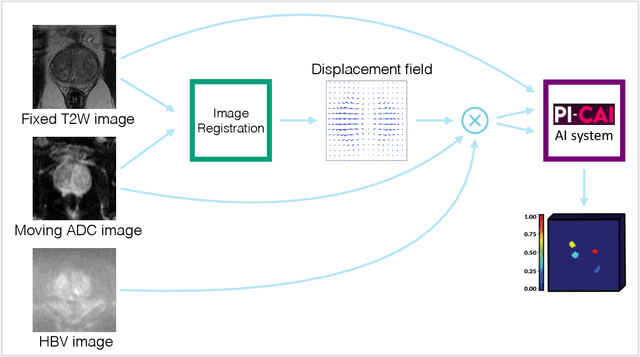
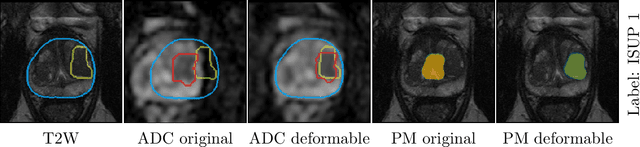
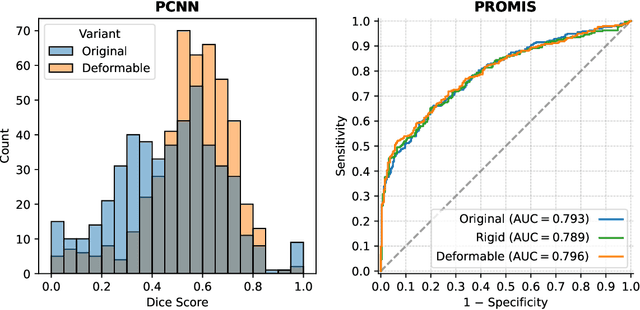
Abstract:The PI-CAI (Prostate Imaging: Cancer AI) challenge led to expert-level diagnostic algorithms for clinically significant prostate cancer detection. The algorithms receive biparametric MRI scans as input, which consist of T2-weighted and diffusion-weighted scans. These scans can be misaligned due to multiple factors in the scanning process. Image registration can alleviate this issue by predicting the deformation between the sequences. We investigate the effect of image registration on the diagnostic performance of AI-based prostate cancer diagnosis. First, the image registration algorithm, developed in MeVisLab, is analyzed using a dataset with paired lesion annotations. Second, the effect on diagnosis is evaluated by comparing case-level cancer diagnosis performance between using the original dataset, rigidly aligned diffusion-weighted scans, or deformably aligned diffusion-weighted scans. Rigid registration showed no improvement. Deformable registration demonstrated a substantial improvement in lesion overlap (+10% median Dice score) and a positive yet non-significant improvement in diagnostic performance (+0.3% AUROC, p=0.18). Our investigation shows that a substantial improvement in lesion alignment does not directly lead to a significant improvement in diagnostic performance. Qualitative analysis indicated that jointly developing image registration methods and diagnostic AI algorithms could enhance diagnostic accuracy and patient outcomes.
Report-Guided Automatic Lesion Annotation for Deep Learning-Based Prostate Cancer Detection in bpMRI
Dec 09, 2021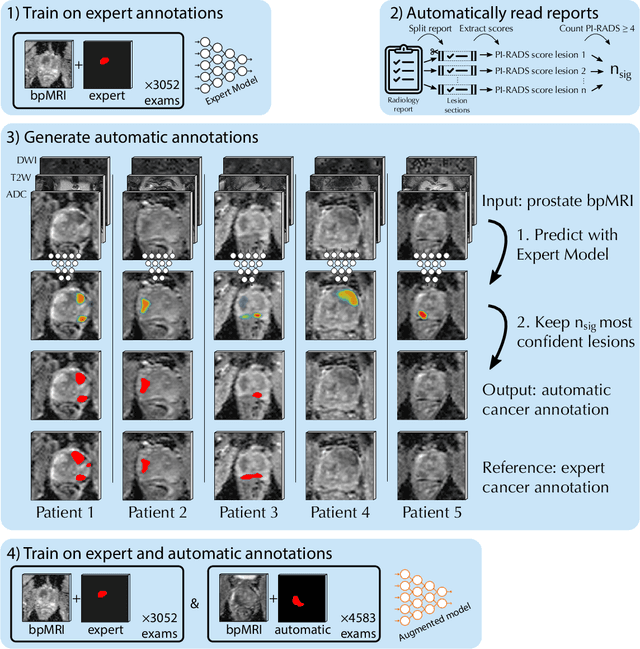

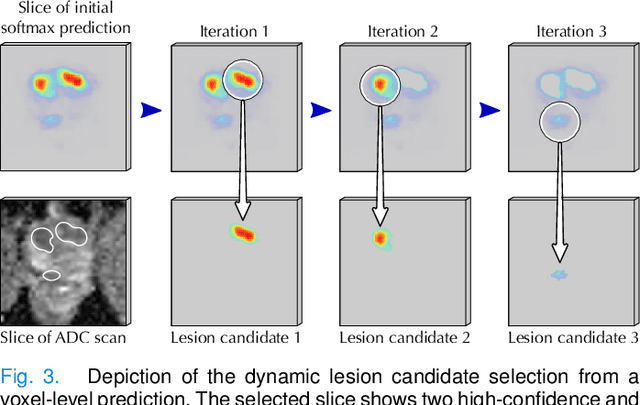
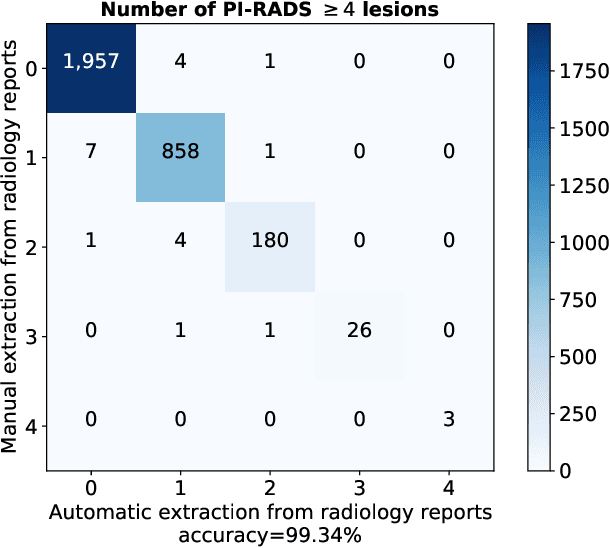
Abstract:Deep learning-based diagnostic performance increases with more annotated data, but manual annotation is a bottleneck in most fields. Experts evaluate diagnostic images during clinical routine, and write their findings in reports. Automatic annotation based on clinical reports could overcome the manual labelling bottleneck. We hypothesise that dense annotations for detection tasks can be generated using model predictions, guided by sparse information from these reports. To demonstrate efficacy, we generated clinically significant prostate cancer (csPCa) annotations, guided by the number of clinically significant findings in the radiology reports. We included 7,756 prostate MRI examinations, of which 3,050 were manually annotated and 4,706 were automatically annotated. We evaluated the automatic annotation quality on the manually annotated subset: our score extraction correctly identified the number of csPCa lesions for $99.3\%$ of the reports and our csPCa segmentation model correctly localised $83.8 \pm 1.1\%$ of the lesions. We evaluated prostate cancer detection performance on 300 exams from an external centre with histopathology-confirmed ground truth. Augmenting the training set with automatically labelled exams improved patient-based diagnostic area under the receiver operating characteristic curve from $88.1\pm 1.1\%$ to $89.8\pm 1.0\%$ ($P = 1.2 \cdot 10^{-4}$) and improved lesion-based sensitivity at one false positive per case from $79.2 \pm 2.8\%$ to $85.4 \pm 1.9\%$ ($P<10^{-4}$), with $mean \pm std.$ over 15 independent runs. This improved performance demonstrates the feasibility of our report-guided automatic annotations. Source code is made publicly available at https://github.com/DIAGNijmegen/Report-Guided-Annotation. Best csPCa detection algorithm is made available at https://grand-challenge.org/algorithms/bpmri-cspca-detection-report-guided-annotations/.
Fully Automatic Deep Learning Framework for Pancreatic Ductal Adenocarcinoma Detection on Computed Tomography
Dec 02, 2021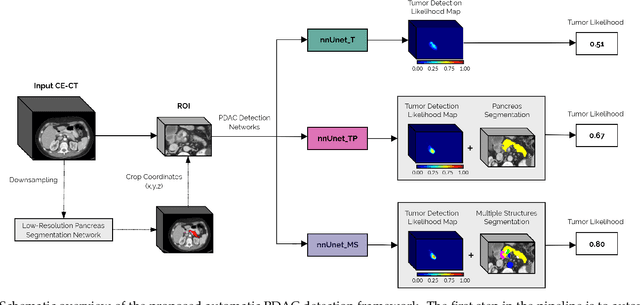

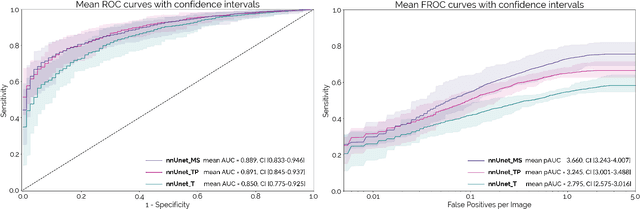
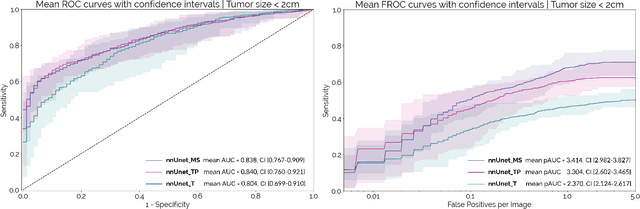
Abstract:Early detection improves prognosis in pancreatic ductal adenocarcinoma (PDAC) but is challenging as lesions are often small and poorly defined on contrast-enhanced computed tomography scans (CE-CT). Deep learning can facilitate PDAC diagnosis, however current models still fail to identify small (<2cm) lesions. In this study, state-of-the-art deep learning models were used to develop an automatic framework for PDAC detection, focusing on small lesions. Additionally, the impact of integrating surrounding anatomy was investigated. CE-CT scans from a cohort of 119 pathology-proven PDAC patients and a cohort of 123 patients without PDAC were used to train a nnUnet for automatic lesion detection and segmentation (nnUnet_T). Two additional nnUnets were trained to investigate the impact of anatomy integration: (1) segmenting the pancreas and tumor (nnUnet_TP), (2) segmenting the pancreas, tumor, and multiple surrounding anatomical structures (nnUnet_MS). An external, publicly available test set was used to compare the performance of the three networks. The nnUnet_MS achieved the best performance, with an area under the receiver operating characteristic curve of 0.91 for the whole test set and 0.88 for tumors <2cm, showing that state-of-the-art deep learning can detect small PDAC and benefits from anatomy information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge