Matin Hosseinzadeh
Uncertainty-Aware Semi-Supervised Learning for Prostate MRI Zonal Segmentation
May 10, 2023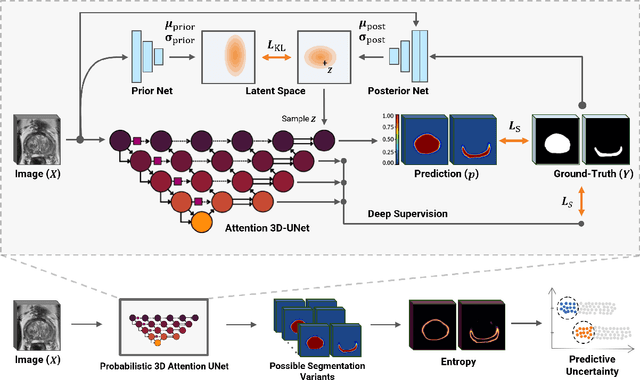
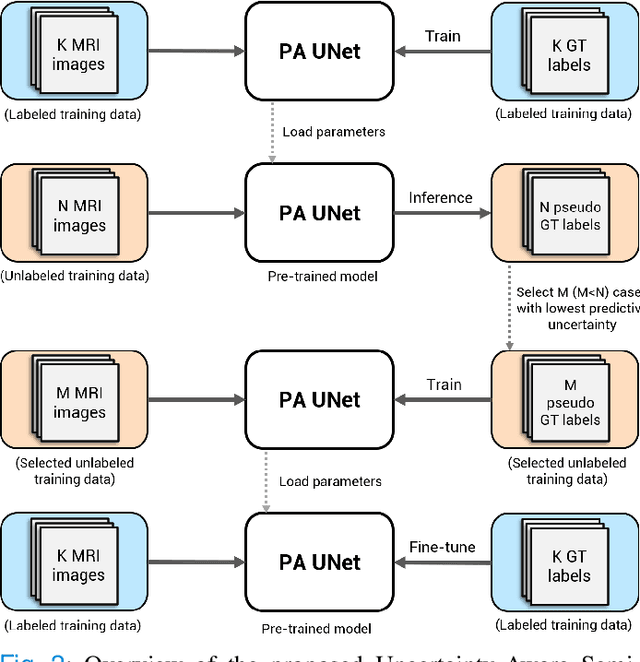
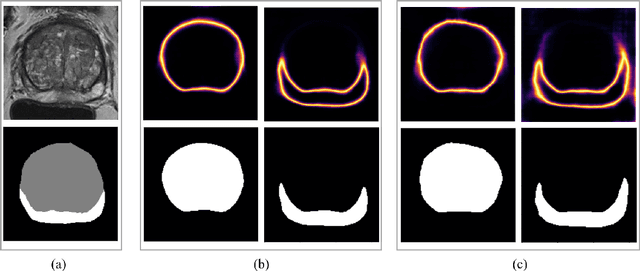
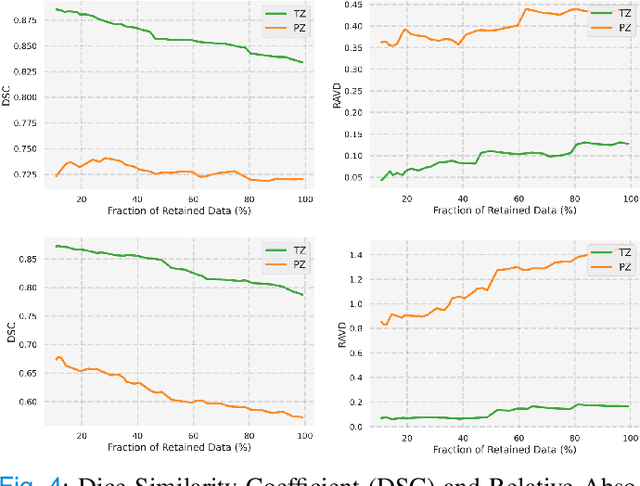
Abstract:Quality of deep convolutional neural network predictions strongly depends on the size of the training dataset and the quality of the annotations. Creating annotations, especially for 3D medical image segmentation, is time-consuming and requires expert knowledge. We propose a novel semi-supervised learning (SSL) approach that requires only a relatively small number of annotations while being able to use the remaining unlabeled data to improve model performance. Our method uses a pseudo-labeling technique that employs recent deep learning uncertainty estimation models. By using the estimated uncertainty, we were able to rank pseudo-labels and automatically select the best pseudo-annotations generated by the supervised model. We applied this to prostate zonal segmentation in T2-weighted MRI scans. Our proposed model outperformed the semi-supervised model in experiments with the ProstateX dataset and an external test set, by leveraging only a subset of unlabeled data rather than the full collection of 4953 cases, our proposed model demonstrated improved performance. The segmentation dice similarity coefficient in the transition zone and peripheral zone increased from 0.835 and 0.727 to 0.852 and 0.751, respectively, for fully supervised model and the uncertainty-aware semi-supervised learning model (USSL). Our USSL model demonstrates the potential to allow deep learning models to be trained on large datasets without requiring full annotation. Our code is available at https://github.com/DIAGNijmegen/prostateMR-USSL.
Report-Guided Automatic Lesion Annotation for Deep Learning-Based Prostate Cancer Detection in bpMRI
Dec 09, 2021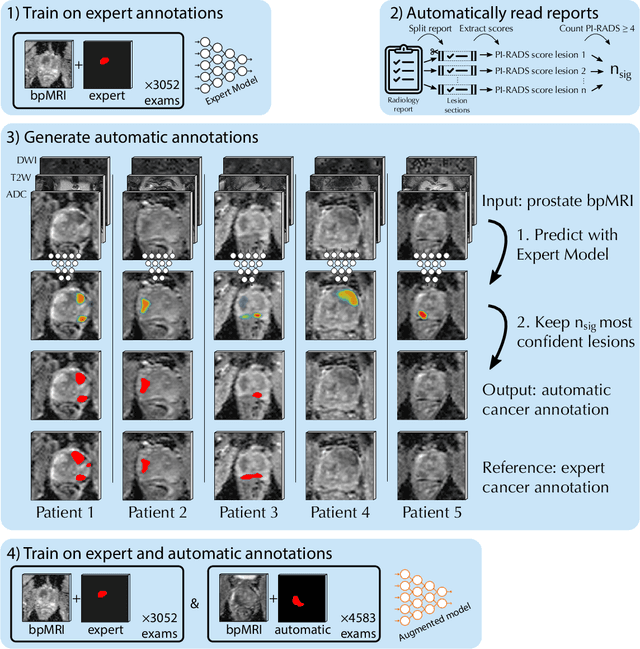

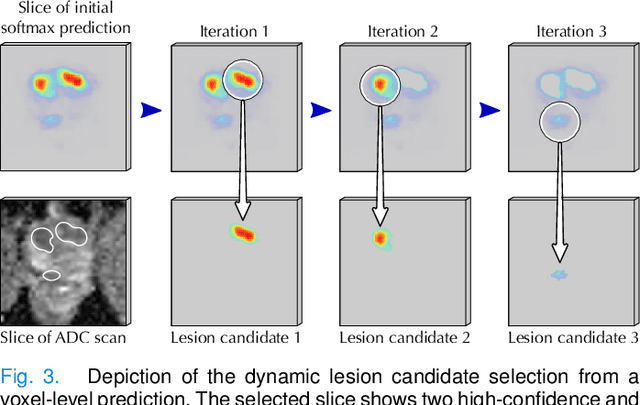
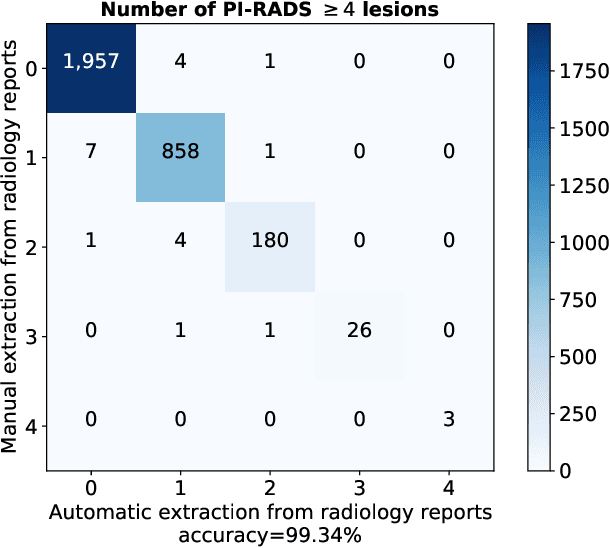
Abstract:Deep learning-based diagnostic performance increases with more annotated data, but manual annotation is a bottleneck in most fields. Experts evaluate diagnostic images during clinical routine, and write their findings in reports. Automatic annotation based on clinical reports could overcome the manual labelling bottleneck. We hypothesise that dense annotations for detection tasks can be generated using model predictions, guided by sparse information from these reports. To demonstrate efficacy, we generated clinically significant prostate cancer (csPCa) annotations, guided by the number of clinically significant findings in the radiology reports. We included 7,756 prostate MRI examinations, of which 3,050 were manually annotated and 4,706 were automatically annotated. We evaluated the automatic annotation quality on the manually annotated subset: our score extraction correctly identified the number of csPCa lesions for $99.3\%$ of the reports and our csPCa segmentation model correctly localised $83.8 \pm 1.1\%$ of the lesions. We evaluated prostate cancer detection performance on 300 exams from an external centre with histopathology-confirmed ground truth. Augmenting the training set with automatically labelled exams improved patient-based diagnostic area under the receiver operating characteristic curve from $88.1\pm 1.1\%$ to $89.8\pm 1.0\%$ ($P = 1.2 \cdot 10^{-4}$) and improved lesion-based sensitivity at one false positive per case from $79.2 \pm 2.8\%$ to $85.4 \pm 1.9\%$ ($P<10^{-4}$), with $mean \pm std.$ over 15 independent runs. This improved performance demonstrates the feasibility of our report-guided automatic annotations. Source code is made publicly available at https://github.com/DIAGNijmegen/Report-Guided-Annotation. Best csPCa detection algorithm is made available at https://grand-challenge.org/algorithms/bpmri-cspca-detection-report-guided-annotations/.
Anatomical and Diagnostic Bayesian Segmentation in Prostate MRI $-$Should Different Clinical Objectives Mandate Different Loss Functions?
Oct 25, 2021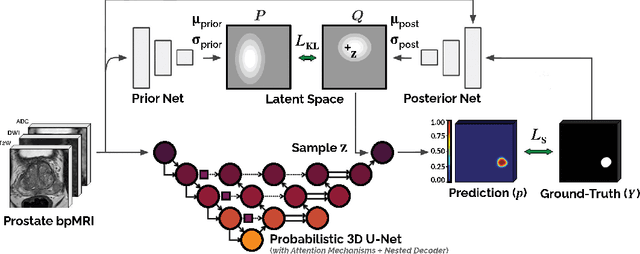
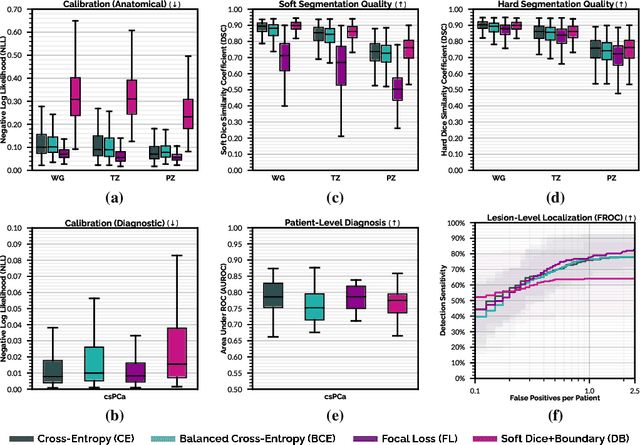
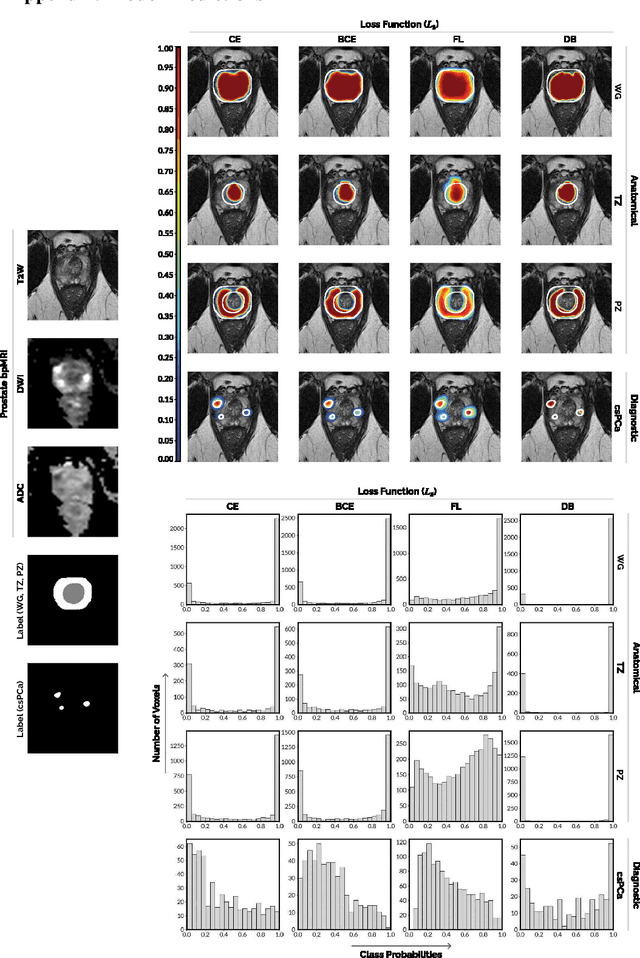
Abstract:We hypothesize that probabilistic voxel-level classification of anatomy and malignancy in prostate MRI, although typically posed as near-identical segmentation tasks via U-Nets, require different loss functions for optimal performance due to inherent differences in their clinical objectives. We investigate distribution, region and boundary-based loss functions for both tasks across 200 patient exams from the publicly-available ProstateX dataset. For evaluation, we conduct a thorough comparative analysis of model predictions and calibration, measured with respect to multi-class volume segmentation of the prostate anatomy (whole-gland, transitional zone, peripheral zone), as well as, patient-level diagnosis and lesion-level detection of clinically significant prostate cancer. Notably, we find that distribution-based loss functions (in particular, focal loss) are well-suited for diagnostic or panoptic segmentation tasks such as lesion detection, primarily due to their implicit property of inducing better calibration. Meanwhile, (with the exception of focal loss) both distribution and region/boundary-based loss functions perform equally well for anatomical or semantic segmentation tasks, such as quantification of organ shape, size and boundaries.
End-to-end Prostate Cancer Detection in bpMRI via 3D CNNs: Effect of Attention Mechanisms, Clinical Priori and Decoupled False Positive Reduction
Jan 28, 2021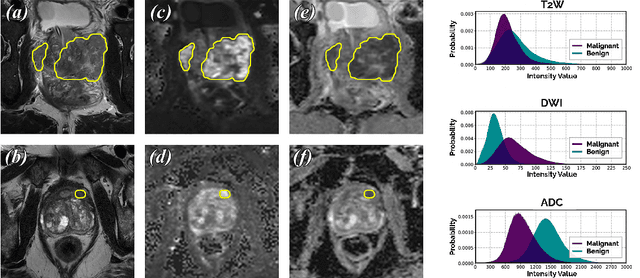
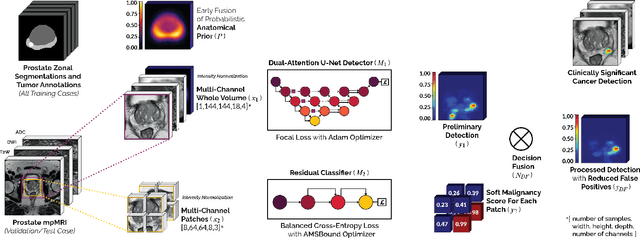
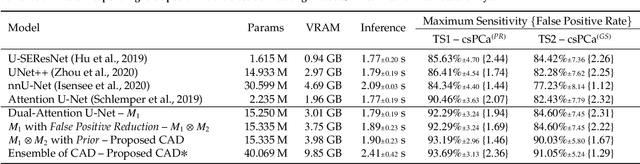
Abstract:We present a novel multi-stage 3D computer-aided detection and diagnosis (CAD) model for automated localization of clinically significant prostate cancer (csPCa) in bi-parametric MR imaging (bpMRI). Deep attention mechanisms drive its detection network, targeting multi-resolution, salient structures and highly discriminative feature dimensions, in order to accurately identify csPCa lesions from indolent cancer and the wide range of benign pathology that can afflict the prostate gland. In parallel, a decoupled residual classifier is used to achieve consistent false positive reduction, without sacrificing high sensitivity or computational efficiency. Furthermore, a probabilistic anatomical prior, which captures the spatial prevalence of csPCa as well as its zonal distinction, is computed and encoded into the CNN architecture to guide model generalization with domain-specific clinical knowledge. For 486 institutional testing scans, the 3D CAD system achieves $83.69\pm5.22\%$ and $93.19\pm2.96\%$ detection sensitivity at 0.50 and 1.46 false positive(s) per patient, respectively, along with $0.882$ AUROC in patient-based diagnosis $-$significantly outperforming four state-of-the-art baseline architectures (U-SEResNet, UNet++, nnU-Net, Attention U-Net) from recent literature. For 296 external testing scans, the ensembled CAD system shares moderate agreement with a consensus of expert radiologists ($76.69\%$; $kappa=0.511$) and independent pathologists ($81.08\%$; $kappa=0.559$); demonstrating strong generalization to histologically-confirmed malignancies, despite using 1950 training-validation cases with radiologically-estimated annotations only.
Encoding Clinical Priori in 3D Convolutional Neural Networks for Prostate Cancer Detection in bpMRI
Nov 03, 2020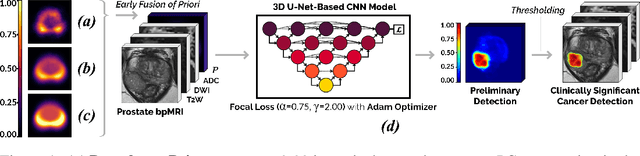
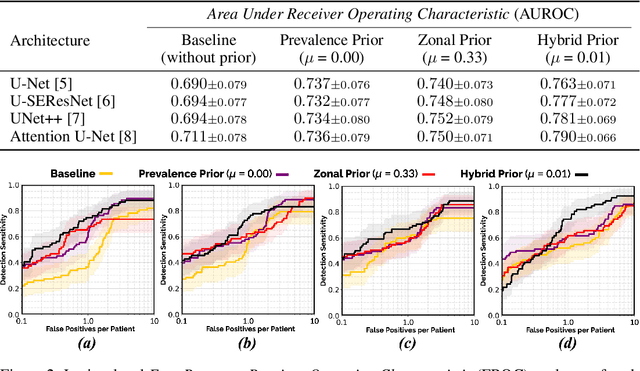
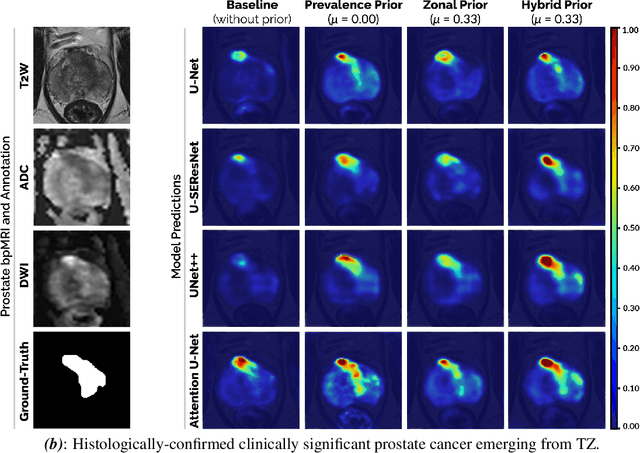
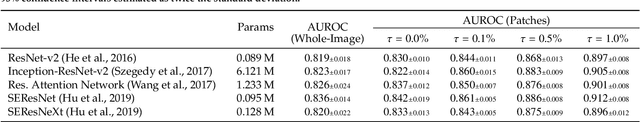
Abstract:We hypothesize that anatomical priors can be viable mediums to infuse domain-specific clinical knowledge into state-of-the-art convolutional neural networks (CNN) based on the U-Net architecture. We introduce a probabilistic population prior which captures the spatial prevalence and zonal distinction of clinically significant prostate cancer (csPCa), in order to improve its computer-aided detection (CAD) in bi-parametric MR imaging (bpMRI). To evaluate performance, we train 3D adaptations of the U-Net, U-SEResNet, UNet++ and Attention U-Net using 800 institutional training-validation scans, paired with radiologically-estimated annotations and our computed prior. For 200 independent testing bpMRI scans with histologically-confirmed delineations of csPCa, our proposed method of encoding clinical priori demonstrates a strong ability to improve patient-based diagnosis (upto 8.70% increase in AUROC) and lesion-level detection (average increase of 1.08 pAUC between 0.1-1.0 false positive per patient) across all four architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge