Tobias Penzkofer
Volumetric Reconstruction of Prostatectomy Specimens from Histology
Nov 29, 2024



Abstract:Surgical treatment for prostate cancer often involves organ removal, i.e., prostatectomy. Pathology reports on these specimens convey treatment-relevant information. Beyond these reports, the diagnostic process generates extensive and complex information that is difficult to represent in reports, although it is of significant interest to the other medical specialties involved. 3D tissue reconstruction would allow for better spatial visualization, as well as combinations with other imaging modalities. Existing approaches in this area have proven labor-intensive and challenging to integrate into clinical workflows. 3D-SLIVER provides a simplified solution, implemented as an open-source 3DSlicer extension. We outline three specific real-world scenarios to illustrate its potential to improve transparency in diagnostic workflows and contribute to multi-modal research endeavors. Implementing the 3D reconstruction process involved four sub-modules of 3D-SLIVER: digitization of slicing protocol, virtual slicing of arbitrary 3D models based on that protocol, registration of slides with virtual slices using the Coherent Point Drift algorithm, and 3D reconstruction of registered information using convex hulls, Gaussian splatter and linear extrusion. Three use cases to employ 3D-SLIVER are presented: a low-effort approach to pathology workflow integration and two research-related use cases illustrating how to perform retrospective evaluations of PI-RADS predictions and statistically model 3D distributions of morphological patterns. 3D-SLIVER allows for improved interdisciplinary communication among specialties. It is designed for simplicity in application, allowing for flexible integration into various workflows and use cases. Here we focused on the clinical care of prostate cancer patients, but future possibilities are extensive with other neoplasms and in education and research.
Deep Learning-based Unsupervised Domain Adaptation via a Unified Model for Prostate Lesion Detection Using Multisite Bi-parametric MRI Datasets
Aug 08, 2024


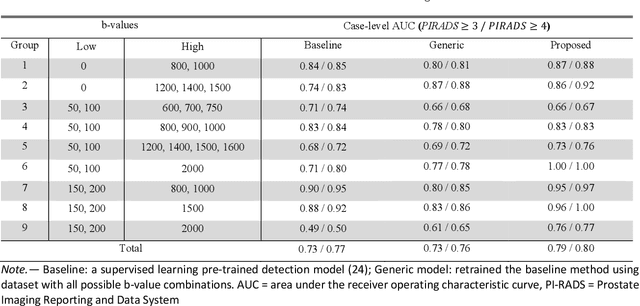
Abstract:Our hypothesis is that UDA using diffusion-weighted images, generated with a unified model, offers a promising and reliable strategy for enhancing the performance of supervised learning models in multi-site prostate lesion detection, especially when various b-values are present. This retrospective study included data from 5,150 patients (14,191 samples) collected across nine different imaging centers. A novel UDA method using a unified generative model was developed for multi-site PCa detection. This method translates diffusion-weighted imaging (DWI) acquisitions, including apparent diffusion coefficient (ADC) and individual DW images acquired using various b-values, to align with the style of images acquired using b-values recommended by Prostate Imaging Reporting and Data System (PI-RADS) guidelines. The generated ADC and DW images replace the original images for PCa detection. An independent set of 1,692 test cases (2,393 samples) was used for evaluation. The area under the receiver operating characteristic curve (AUC) was used as the primary metric, and statistical analysis was performed via bootstrapping. For all test cases, the AUC values for baseline SL and UDA methods were 0.73 and 0.79 (p<.001), respectively, for PI-RADS>=3, and 0.77 and 0.80 (p<.001) for PI-RADS>=4 PCa lesions. In the 361 test cases under the most unfavorable image acquisition setting, the AUC values for baseline SL and UDA were 0.49 and 0.76 (p<.001) for PI-RADS>=3, and 0.50 and 0.77 (p<.001) for PI-RADS>=4 PCa lesions. The results indicate the proposed UDA with generated images improved the performance of SL methods in multi-site PCa lesion detection across datasets with various b values, especially for images acquired with significant deviations from the PI-RADS recommended DWI protocol (e.g. with an extremely high b-value).
Real-World Federated Learning in Radiology: Hurdles to overcome and Benefits to gain
May 15, 2024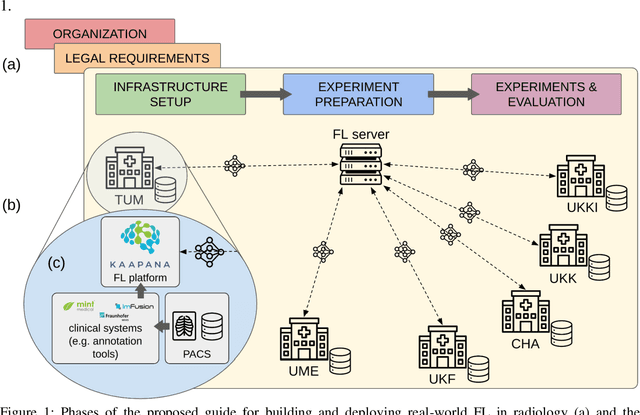



Abstract:Objective: Federated Learning (FL) enables collaborative model training while keeping data locally. Currently, most FL studies in radiology are conducted in simulated environments due to numerous hurdles impeding its translation into practice. The few existing real-world FL initiatives rarely communicate specific measures taken to overcome these hurdles, leaving behind a significant knowledge gap. Minding efforts to implement real-world FL, there is a notable lack of comprehensive assessment comparing FL to less complex alternatives. Materials & Methods: We extensively reviewed FL literature, categorizing insights along with our findings according to their nature and phase while establishing a FL initiative, summarized to a comprehensive guide. We developed our own FL infrastructure within the German Radiological Cooperative Network (RACOON) and demonstrated its functionality by training FL models on lung pathology segmentation tasks across six university hospitals. We extensively evaluated FL against less complex alternatives in three distinct evaluation scenarios. Results: The proposed guide outlines essential steps, identified hurdles, and proposed solutions for establishing successful FL initiatives conducting real-world experiments. Our experimental results show that FL outperforms less complex alternatives in all evaluation scenarios, justifying the effort required to translate FL into real-world applications. Discussion & Conclusion: Our proposed guide aims to aid future FL researchers in circumventing pitfalls and accelerating translation of FL into radiological applications. Our results underscore the value of efforts needed to translate FL into real-world applications by demonstrating advantageous performance over alternatives, and emphasize the importance of strategic organization, robust management of distributed data and infrastructure in real-world settings.
Leveraging Foundation Models for Content-Based Medical Image Retrieval in Radiology
Mar 11, 2024Abstract:Content-based image retrieval (CBIR) has the potential to significantly improve diagnostic aid and medical research in radiology. Current CBIR systems face limitations due to their specialization to certain pathologies, limiting their utility. In response, we propose using vision foundation models as powerful and versatile off-the-shelf feature extractors for content-based medical image retrieval. By benchmarking these models on a comprehensive dataset of 1.6 million 2D radiological images spanning four modalities and 161 pathologies, we identify weakly-supervised models as superior, achieving a P@1 of up to 0.594. This performance not only competes with a specialized model but does so without the need for fine-tuning. Our analysis further explores the challenges in retrieving pathological versus anatomical structures, indicating that accurate retrieval of pathological features presents greater difficulty. Despite these challenges, our research underscores the vast potential of foundation models for CBIR in radiology, proposing a shift towards versatile, general-purpose medical image retrieval systems that do not require specific tuning.
Efficient Large Scale Medical Image Dataset Preparation for Machine Learning Applications
Sep 29, 2023Abstract:In the rapidly evolving field of medical imaging, machine learning algorithms have become indispensable for enhancing diagnostic accuracy. However, the effectiveness of these algorithms is contingent upon the availability and organization of high-quality medical imaging datasets. Traditional Digital Imaging and Communications in Medicine (DICOM) data management systems are inadequate for handling the scale and complexity of data required to be facilitated in machine learning algorithms. This paper introduces an innovative data curation tool, developed as part of the Kaapana open-source toolkit, aimed at streamlining the organization, management, and processing of large-scale medical imaging datasets. The tool is specifically tailored to meet the needs of radiologists and machine learning researchers. It incorporates advanced search, auto-annotation and efficient tagging functionalities for improved data curation. Additionally, the tool facilitates quality control and review, enabling researchers to validate image and segmentation quality in large datasets. It also plays a critical role in uncovering potential biases in datasets by aggregating and visualizing metadata, which is essential for developing robust machine learning models. Furthermore, Kaapana is integrated within the Radiological Cooperative Network (RACOON), a pioneering initiative aimed at creating a comprehensive national infrastructure for the aggregation, transmission, and consolidation of radiological data across all university clinics throughout Germany. A supplementary video showcasing the tool's functionalities can be accessed at https://bit.ly/MICCAI-DEMI2023.
Current State of Community-Driven Radiological AI Deployment in Medical Imaging
Dec 29, 2022



Abstract:Artificial Intelligence (AI) has become commonplace to solve routine everyday tasks. Because of the exponential growth in medical imaging data volume and complexity, the workload on radiologists is steadily increasing. We project that the gap between the number of imaging exams and the number of expert radiologist readers required to cover this increase will continue to expand, consequently introducing a demand for AI-based tools that improve the efficiency with which radiologists can comfortably interpret these exams. AI has been shown to improve efficiency in medical-image generation, processing, and interpretation, and a variety of such AI models have been developed across research labs worldwide. However, very few of these, if any, find their way into routine clinical use, a discrepancy that reflects the divide between AI research and successful AI translation. To address the barrier to clinical deployment, we have formed MONAI Consortium, an open-source community which is building standards for AI deployment in healthcare institutions, and developing tools and infrastructure to facilitate their implementation. This report represents several years of weekly discussions and hands-on problem solving experience by groups of industry experts and clinicians in the MONAI Consortium. We identify barriers between AI-model development in research labs and subsequent clinical deployment and propose solutions. Our report provides guidance on processes which take an imaging AI model from development to clinical implementation in a healthcare institution. We discuss various AI integration points in a clinical Radiology workflow. We also present a taxonomy of Radiology AI use-cases. Through this report, we intend to educate the stakeholders in healthcare and AI (AI researchers, radiologists, imaging informaticists, and regulators) about cross-disciplinary challenges and possible solutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge