Paul F. Jäger
Finally Outshining the Random Baseline: A Simple and Effective Solution for Active Learning in 3D Biomedical Imaging
Jan 20, 2026Abstract:Active learning (AL) has the potential to drastically reduce annotation costs in 3D biomedical image segmentation, where expert labeling of volumetric data is both time-consuming and expensive. Yet, existing AL methods are unable to consistently outperform improved random sampling baselines adapted to 3D data, leaving the field without a reliable solution. We introduce Class-stratified Scheduled Power Predictive Entropy (ClaSP PE), a simple and effective query strategy that addresses two key limitations of standard uncertainty-based AL methods: class imbalance and redundancy in early selections. ClaSP PE combines class-stratified querying to ensure coverage of underrepresented structures and log-scale power noising with a decaying schedule to enforce query diversity in early-stage AL and encourage exploitation later. In our evaluation on 24 experimental settings using four 3D biomedical datasets within the comprehensive nnActive benchmark, ClaSP PE is the only method that generally outperforms improved random baselines in terms of both segmentation quality with statistically significant gains, whilst remaining annotation efficient. Furthermore, we explicitly simulate the real-world application by testing our method on four previously unseen datasets without manual adaptation, where all experiment parameters are set according to predefined guidelines. The results confirm that ClaSP PE robustly generalizes to novel tasks without requiring dataset-specific tuning. Within the nnActive framework, we present compelling evidence that an AL method can consistently outperform random baselines adapted to 3D segmentation, in terms of both performance and annotation efficiency in a realistic, close-to-production scenario. Our open-source implementation and clear deployment guidelines make it readily applicable in practice. Code is at https://github.com/MIC-DKFZ/nnActive.
Revisiting MAE pre-training for 3D medical image segmentation
Oct 30, 2024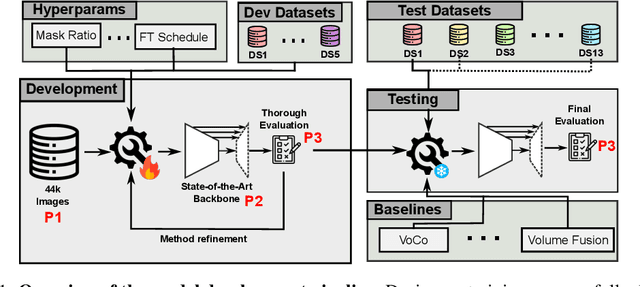
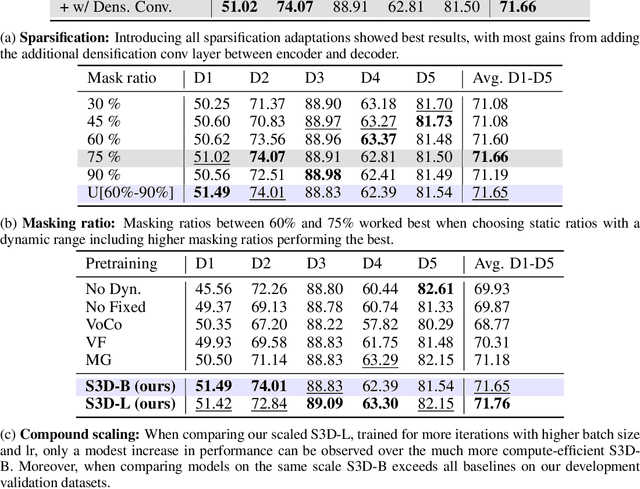
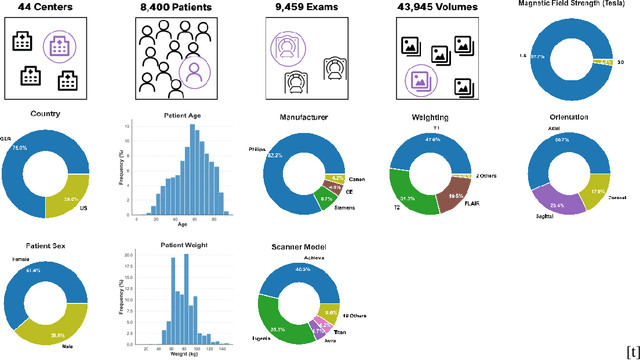
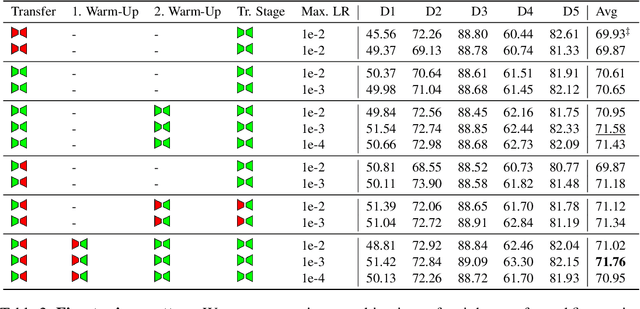
Abstract:Self-Supervised Learning (SSL) presents an exciting opportunity to unlock the potential of vast, untapped clinical datasets, for various downstream applications that suffer from the scarcity of labeled data. While SSL has revolutionized fields like natural language processing and computer vision, their adoption in 3D medical image computing has been limited by three key pitfalls: Small pre-training dataset sizes, architectures inadequate for 3D medical image analysis, and insufficient evaluation practices. We address these issues by i) leveraging a large-scale dataset of 44k 3D brain MRI volumes and ii) using a Residual Encoder U-Net architecture within the state-of-the-art nnU-Net framework. iii) A robust development framework, incorporating 5 development and 8 testing brain MRI segmentation datasets, allowed performance-driven design decisions to optimize the simple concept of Masked Auto Encoders (MAEs) for 3D CNNs. The resulting model not only surpasses previous SSL methods but also outperforms the strong nnU-Net baseline by an average of approximately 3 Dice points. Furthermore, our model demonstrates exceptional stability, achieving the highest average rank of 2 out of 7 methods, compared to the second-best method's mean rank of 3.
Confidence intervals uncovered: Are we ready for real-world medical imaging AI?
Sep 27, 2024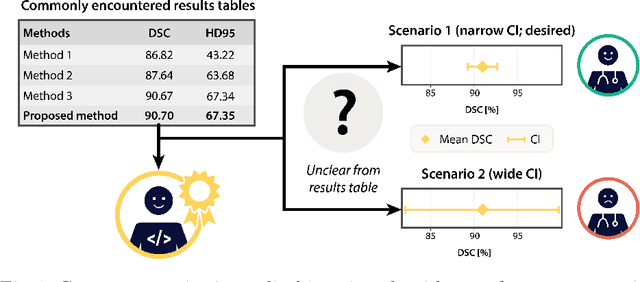
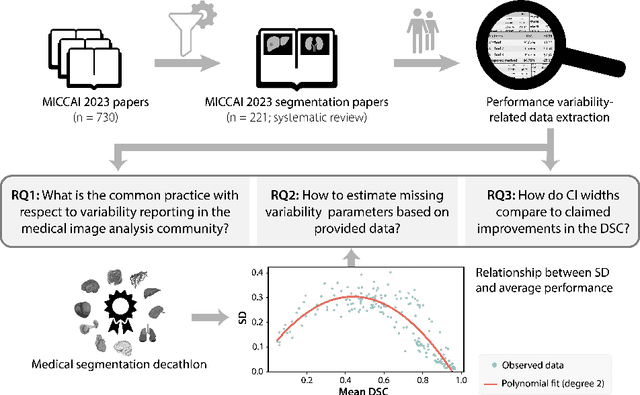
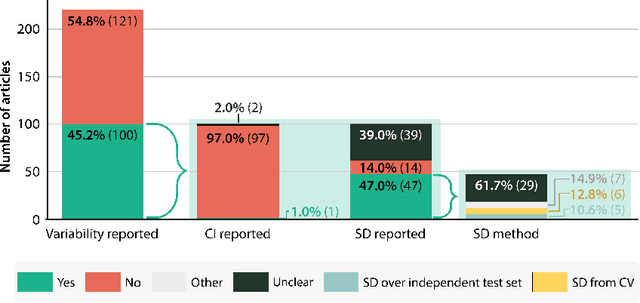

Abstract:Medical imaging is spearheading the AI transformation of healthcare. Performance reporting is key to determine which methods should be translated into clinical practice. Frequently, broad conclusions are simply derived from mean performance values. In this paper, we argue that this common practice is often a misleading simplification as it ignores performance variability. Our contribution is threefold. (1) Analyzing all MICCAI segmentation papers (n = 221) published in 2023, we first observe that more than 50% of papers do not assess performance variability at all. Moreover, only one (0.5%) paper reported confidence intervals (CIs) for model performance. (2) To address the reporting bottleneck, we show that the unreported standard deviation (SD) in segmentation papers can be approximated by a second-order polynomial function of the mean Dice similarity coefficient (DSC). Based on external validation data from 56 previous MICCAI challenges, we demonstrate that this approximation can accurately reconstruct the CI of a method using information provided in publications. (3) Finally, we reconstructed 95% CIs around the mean DSC of MICCAI 2023 segmentation papers. The median CI width was 0.03 which is three times larger than the median performance gap between the first and second ranked method. For more than 60% of papers, the mean performance of the second-ranked method was within the CI of the first-ranked method. We conclude that current publications typically do not provide sufficient evidence to support which models could potentially be translated into clinical practice.
Navigating the Maze of Explainable AI: A Systematic Approach to Evaluating Methods and Metrics
Sep 25, 2024Abstract:Explainable AI (XAI) is a rapidly growing domain with a myriad of proposed methods as well as metrics aiming to evaluate their efficacy. However, current studies are often of limited scope, examining only a handful of XAI methods and ignoring underlying design parameters for performance, such as the model architecture or the nature of input data. Moreover, they often rely on one or a few metrics and neglect thorough validation, increasing the risk of selection bias and ignoring discrepancies among metrics. These shortcomings leave practitioners confused about which method to choose for their problem. In response, we introduce LATEC, a large-scale benchmark that critically evaluates 17 prominent XAI methods using 20 distinct metrics. We systematically incorporate vital design parameters like varied architectures and diverse input modalities, resulting in 7,560 examined combinations. Through LATEC, we showcase the high risk of conflicting metrics leading to unreliable rankings and consequently propose a more robust evaluation scheme. Further, we comprehensively evaluate various XAI methods to assist practitioners in selecting appropriate methods aligning with their needs. Curiously, the emerging top-performing method, Expected Gradients, is not examined in any relevant related study. LATEC reinforces its role in future XAI research by publicly releasing all 326k saliency maps and 378k metric scores as a (meta-)evaluation dataset.
Visual Prompt Engineering for Medical Vision Language Models in Radiology
Aug 28, 2024



Abstract:Medical image classification in radiology faces significant challenges, particularly in generalizing to unseen pathologies. In contrast, CLIP offers a promising solution by leveraging multimodal learning to improve zero-shot classification performance. However, in the medical domain, lesions can be small and might not be well represented in the embedding space. Therefore, in this paper, we explore the potential of visual prompt engineering to enhance the capabilities of Vision Language Models (VLMs) in radiology. Leveraging BiomedCLIP, trained on extensive biomedical image-text pairs, we investigate the impact of embedding visual markers directly within radiological images to guide the model's attention to critical regions. Our evaluation on the JSRT dataset, focusing on lung nodule malignancy classification, demonstrates that incorporating visual prompts $\unicode{x2013}$ such as arrows, circles, and contours $\unicode{x2013}$ significantly improves classification metrics including AUROC, AUPRC, F1 score, and accuracy. Moreover, the study provides attention maps, showcasing enhanced model interpretability and focus on clinically relevant areas. These findings underscore the efficacy of visual prompt engineering as a straightforward yet powerful approach to advance VLM performance in medical image analysis.
Comparative Benchmarking of Failure Detection Methods in Medical Image Segmentation: Unveiling the Role of Confidence Aggregation
Jun 05, 2024



Abstract:Semantic segmentation is an essential component of medical image analysis research, with recent deep learning algorithms offering out-of-the-box applicability across diverse datasets. Despite these advancements, segmentation failures remain a significant concern for real-world clinical applications, necessitating reliable detection mechanisms. This paper introduces a comprehensive benchmarking framework aimed at evaluating failure detection methodologies within medical image segmentation. Through our analysis, we identify the strengths and limitations of current failure detection metrics, advocating for the risk-coverage analysis as a holistic evaluation approach. Utilizing a collective dataset comprising five public 3D medical image collections, we assess the efficacy of various failure detection strategies under realistic test-time distribution shifts. Our findings highlight the importance of pixel confidence aggregation and we observe superior performance of the pairwise Dice score (Roy et al., 2019) between ensemble predictions, positioning it as a simple and robust baseline for failure detection in medical image segmentation. To promote ongoing research, we make the benchmarking framework available to the community.
Embarrassingly Simple Scribble Supervision for 3D Medical Segmentation
Mar 19, 2024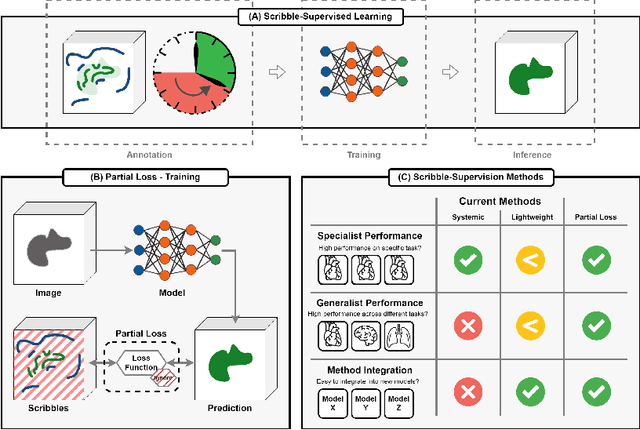
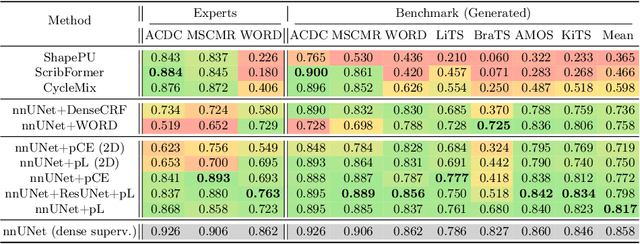
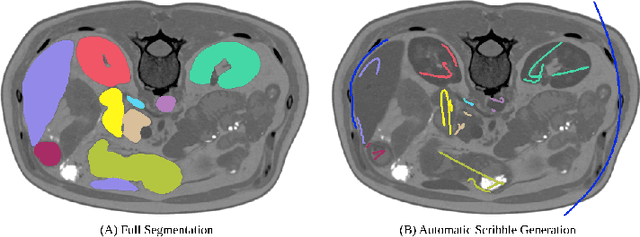
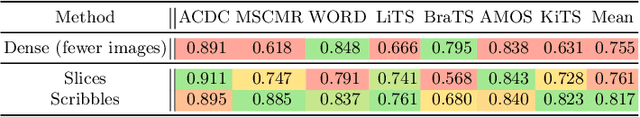
Abstract:Traditionally, segmentation algorithms require dense annotations for training, demanding significant annotation efforts, particularly within the 3D medical imaging field. Scribble-supervised learning emerges as a possible solution to this challenge, promising a reduction in annotation efforts when creating large-scale datasets. Recently, a plethora of methods for optimized learning from scribbles have been proposed, but have so far failed to position scribble annotation as a beneficial alternative. We relate this shortcoming to two major issues: 1) the complex nature of many methods which deeply ties them to the underlying segmentation model, thus preventing a migration to more powerful state-of-the-art models as the field progresses and 2) the lack of a systematic evaluation to validate consistent performance across the broader medical domain, resulting in a lack of trust when applying these methods to new segmentation problems. To address these issues, we propose a comprehensive scribble supervision benchmark consisting of seven datasets covering a diverse set of anatomies and pathologies imaged with varying modalities. We furthermore propose the systematic use of partial losses, i.e. losses that are only computed on annotated voxels. Contrary to most existing methods, these losses can be seamlessly integrated into state-of-the-art segmentation methods, enabling them to learn from scribble annotations while preserving their original loss formulations. Our evaluation using nnU-Net reveals that while most existing methods suffer from a lack of generalization, the proposed approach consistently delivers state-of-the-art performance. Thanks to its simplicity, our approach presents an embarrassingly simple yet effective solution to the challenges of scribble supervision. Source code as well as our extensive scribble benchmarking suite will be made publicly available upon publication.
Leveraging Foundation Models for Content-Based Medical Image Retrieval in Radiology
Mar 11, 2024Abstract:Content-based image retrieval (CBIR) has the potential to significantly improve diagnostic aid and medical research in radiology. Current CBIR systems face limitations due to their specialization to certain pathologies, limiting their utility. In response, we propose using vision foundation models as powerful and versatile off-the-shelf feature extractors for content-based medical image retrieval. By benchmarking these models on a comprehensive dataset of 1.6 million 2D radiological images spanning four modalities and 161 pathologies, we identify weakly-supervised models as superior, achieving a P@1 of up to 0.594. This performance not only competes with a specialized model but does so without the need for fine-tuning. Our analysis further explores the challenges in retrieving pathological versus anatomical structures, indicating that accurate retrieval of pathological features presents greater difficulty. Despite these challenges, our research underscores the vast potential of foundation models for CBIR in radiology, proposing a shift towards versatile, general-purpose medical image retrieval systems that do not require specific tuning.
Deep Interactive Segmentation of Medical Images: A Systematic Review and Taxonomy
Nov 23, 2023Abstract:Interactive segmentation is a crucial research area in medical image analysis aiming to boost the efficiency of costly annotations by incorporating human feedback. This feedback takes the form of clicks, scribbles, or masks and allows for iterative refinement of the model output so as to efficiently guide the system towards the desired behavior. In recent years, deep learning-based approaches have propelled results to a new level causing a rapid growth in the field with 121 methods proposed in the medical imaging domain alone. In this review, we provide a structured overview of this emerging field featuring a comprehensive taxonomy, a systematic review of existing methods, and an in-depth analysis of current practices. Based on these contributions, we discuss the challenges and opportunities in the field. For instance, we find that there is a severe lack of comparison across methods which needs to be tackled by standardized baselines and benchmarks.
Why is the winner the best?
Mar 30, 2023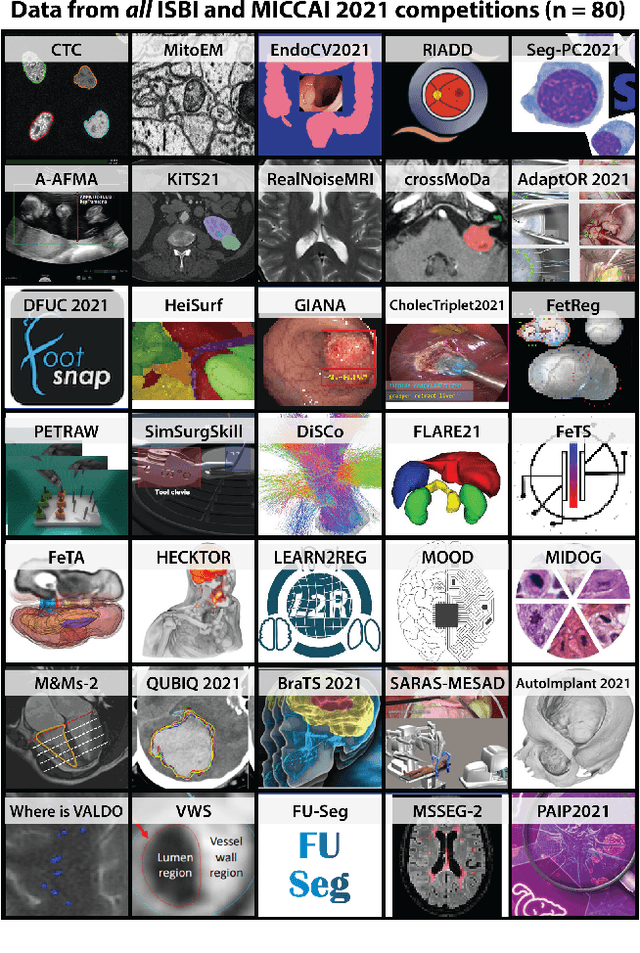
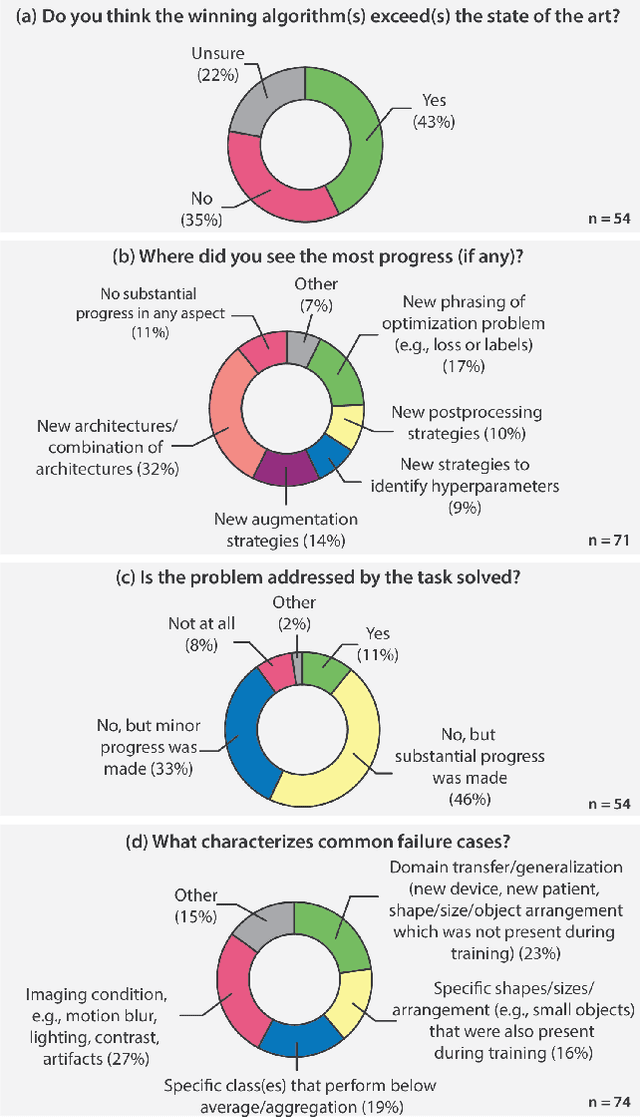
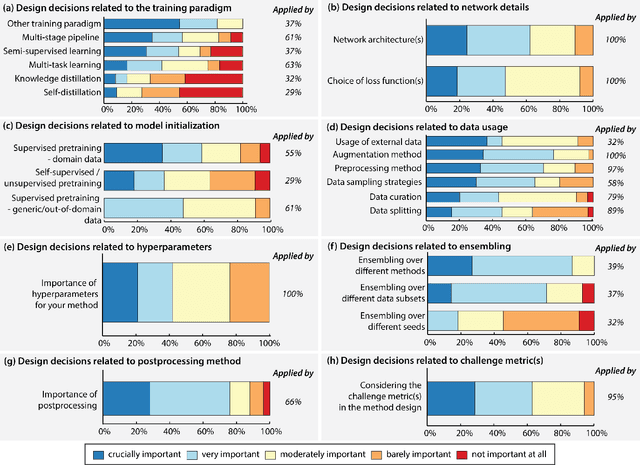
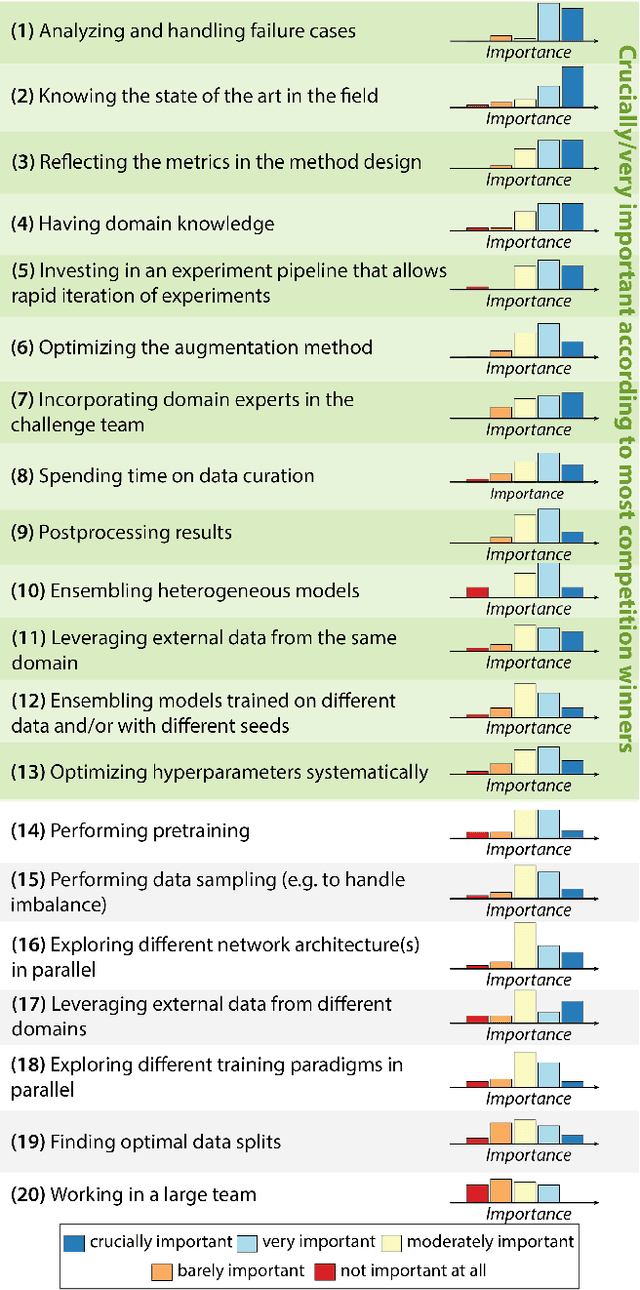
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge