Maëlys Solal
ARAMIS
Medical Imaging AI Competitions Lack Fairness
Dec 19, 2025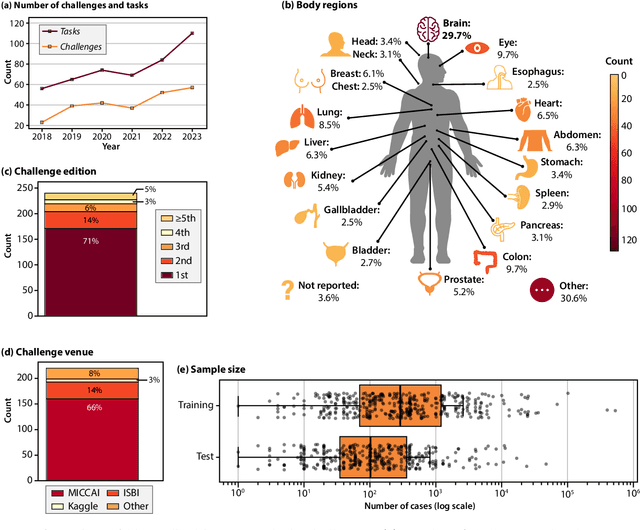
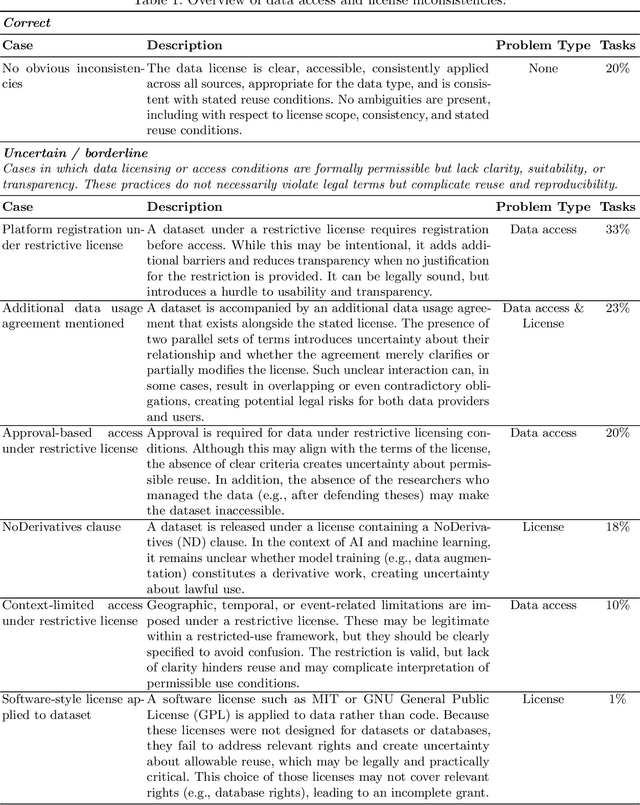
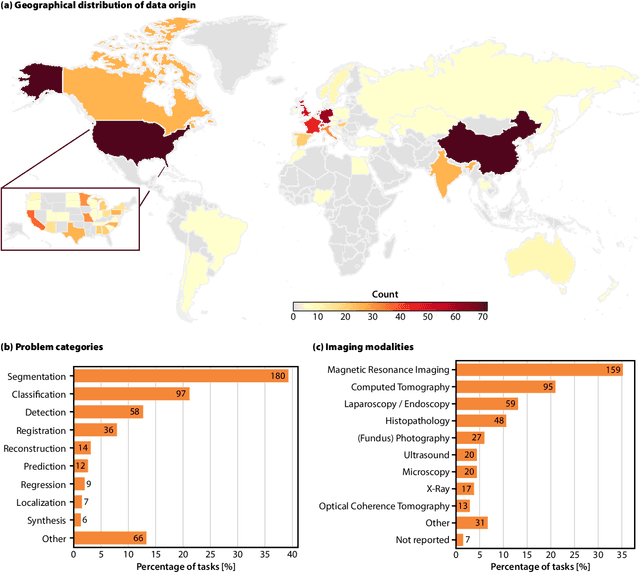
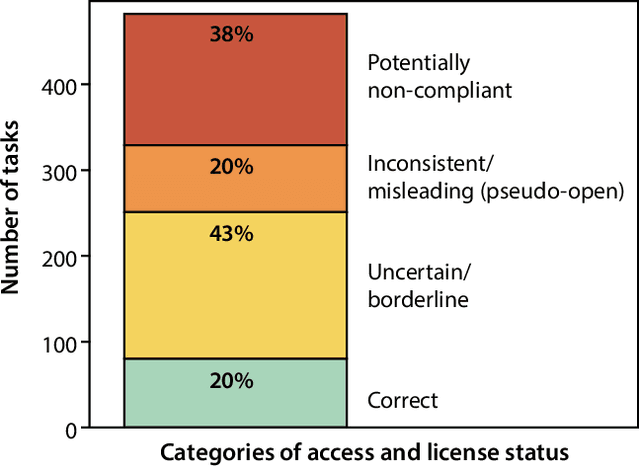
Abstract:Benchmarking competitions are central to the development of artificial intelligence (AI) in medical imaging, defining performance standards and shaping methodological progress. However, it remains unclear whether these benchmarks provide data that are sufficiently representative, accessible, and reusable to support clinically meaningful AI. In this work, we assess fairness along two complementary dimensions: (1) whether challenge datasets are representative of real-world clinical diversity, and (2) whether they are accessible and legally reusable in line with the FAIR principles. To address this question, we conducted a large-scale systematic study of 241 biomedical image analysis challenges comprising 458 tasks across 19 imaging modalities. Our findings show substantial biases in dataset composition, including geographic location, modality-, and problem type-related biases, indicating that current benchmarks do not adequately reflect real-world clinical diversity. Despite their widespread influence, challenge datasets were frequently constrained by restrictive or ambiguous access conditions, inconsistent or non-compliant licensing practices, and incomplete documentation, limiting reproducibility and long-term reuse. Together, these shortcomings expose foundational fairness limitations in our benchmarking ecosystem and highlight a disconnect between leaderboard success and clinical relevance.
False Promises in Medical Imaging AI? Assessing Validity of Outperformance Claims
May 07, 2025Abstract:Performance comparisons are fundamental in medical imaging Artificial Intelligence (AI) research, often driving claims of superiority based on relative improvements in common performance metrics. However, such claims frequently rely solely on empirical mean performance. In this paper, we investigate whether newly proposed methods genuinely outperform the state of the art by analyzing a representative cohort of medical imaging papers. We quantify the probability of false claims based on a Bayesian approach that leverages reported results alongside empirically estimated model congruence to estimate whether the relative ranking of methods is likely to have occurred by chance. According to our results, the majority (>80%) of papers claims outperformance when introducing a new method. Our analysis further revealed a high probability (>5%) of false outperformance claims in 86% of classification papers and 53% of segmentation papers. These findings highlight a critical flaw in current benchmarking practices: claims of outperformance in medical imaging AI are frequently unsubstantiated, posing a risk of misdirecting future research efforts.
Confidence intervals uncovered: Are we ready for real-world medical imaging AI?
Sep 27, 2024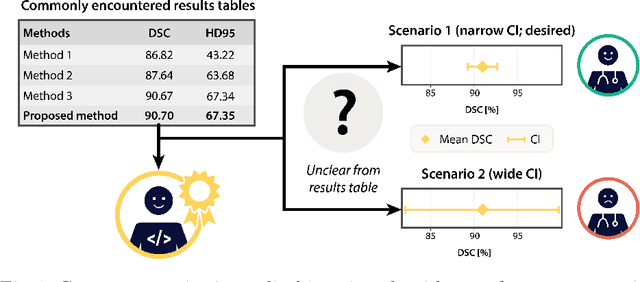
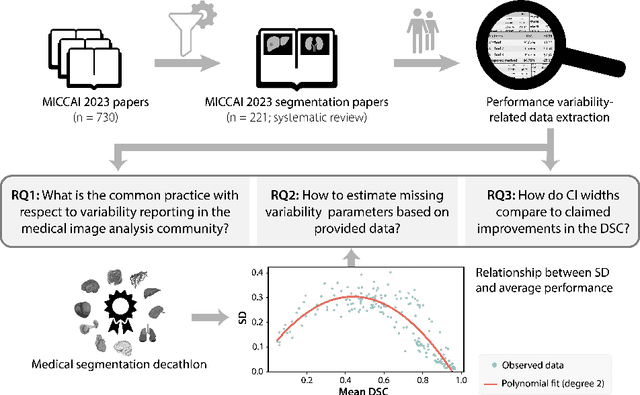
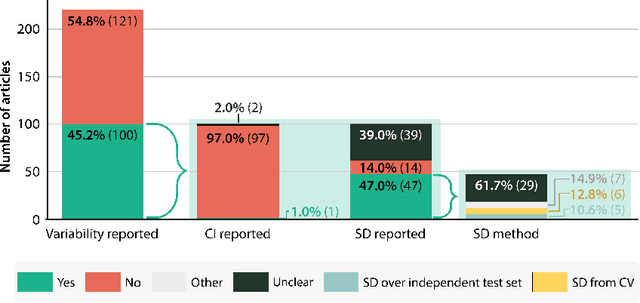

Abstract:Medical imaging is spearheading the AI transformation of healthcare. Performance reporting is key to determine which methods should be translated into clinical practice. Frequently, broad conclusions are simply derived from mean performance values. In this paper, we argue that this common practice is often a misleading simplification as it ignores performance variability. Our contribution is threefold. (1) Analyzing all MICCAI segmentation papers (n = 221) published in 2023, we first observe that more than 50% of papers do not assess performance variability at all. Moreover, only one (0.5%) paper reported confidence intervals (CIs) for model performance. (2) To address the reporting bottleneck, we show that the unreported standard deviation (SD) in segmentation papers can be approximated by a second-order polynomial function of the mean Dice similarity coefficient (DSC). Based on external validation data from 56 previous MICCAI challenges, we demonstrate that this approximation can accurately reconstruct the CI of a method using information provided in publications. (3) Finally, we reconstructed 95% CIs around the mean DSC of MICCAI 2023 segmentation papers. The median CI width was 0.03 which is three times larger than the median performance gap between the first and second ranked method. For more than 60% of papers, the mean performance of the second-ranked method was within the CI of the first-ranked method. We conclude that current publications typically do not provide sufficient evidence to support which models could potentially be translated into clinical practice.
Evaluation of pseudo-healthy image reconstruction for anomaly detection with deep generative models: Application to brain FDG PET
Jan 29, 2024Abstract:Over the past years, pseudo-healthy reconstruction for unsupervised anomaly detection has gained in popularity. This approach has the great advantage of not requiring tedious pixel-wise data annotation and offers possibility to generalize to any kind of anomalies, including that corresponding to rare diseases. By training a deep generative model with only images from healthy subjects, the model will learn to reconstruct pseudo-healthy images. This pseudo-healthy reconstruction is then compared to the input to detect and localize anomalies. The evaluation of such methods often relies on a ground truth lesion mask that is available for test data, which may not exist depending on the application. We propose an evaluation procedure based on the simulation of realistic abnormal images to validate pseudo-healthy reconstruction methods when no ground truth is available. This allows us to extensively test generative models on different kinds of anomalies and measuring their performance using the pair of normal and abnormal images corresponding to the same subject. It can be used as a preliminary automatic step to validate the capacity of a generative model to reconstruct pseudo-healthy images, before a more advanced validation step that would require clinician's expertise. We apply this framework to the reconstruction of 3D brain FDG PET using a convolutional variational autoencoder with the aim to detect as early as possible the neurodegeneration markers that are specific to dementia such as Alzheimer's disease.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:003
Leveraging healthy population variability in deep learning unsupervised anomaly detection in brain FDG PET
Nov 20, 2023Abstract:Unsupervised anomaly detection is a popular approach for the analysis of neuroimaging data as it allows to identify a wide variety of anomalies from unlabelled data. It relies on building a subject-specific model of healthy appearance to which a subject's image can be compared to detect anomalies. In the literature, it is common for anomaly detection to rely on analysing the residual image between the subject's image and its pseudo-healthy reconstruction. This approach however has limitations partly due to the pseudo-healthy reconstructions being imperfect and to the lack of natural thresholding mechanism. Our proposed method, inspired by Z-scores, leverages the healthy population variability to overcome these limitations. Our experiments conducted on FDG PET scans from the ADNI database demonstrate the effectiveness of our approach in accurately identifying Alzheimer's disease related anomalies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge