Selen Erkan
False Promises in Medical Imaging AI? Assessing Validity of Outperformance Claims
May 07, 2025Abstract:Performance comparisons are fundamental in medical imaging Artificial Intelligence (AI) research, often driving claims of superiority based on relative improvements in common performance metrics. However, such claims frequently rely solely on empirical mean performance. In this paper, we investigate whether newly proposed methods genuinely outperform the state of the art by analyzing a representative cohort of medical imaging papers. We quantify the probability of false claims based on a Bayesian approach that leverages reported results alongside empirically estimated model congruence to estimate whether the relative ranking of methods is likely to have occurred by chance. According to our results, the majority (>80%) of papers claims outperformance when introducing a new method. Our analysis further revealed a high probability (>5%) of false outperformance claims in 86% of classification papers and 53% of segmentation papers. These findings highlight a critical flaw in current benchmarking practices: claims of outperformance in medical imaging AI are frequently unsubstantiated, posing a risk of misdirecting future research efforts.
SURE-VQA: Systematic Understanding of Robustness Evaluation in Medical VQA Tasks
Nov 29, 2024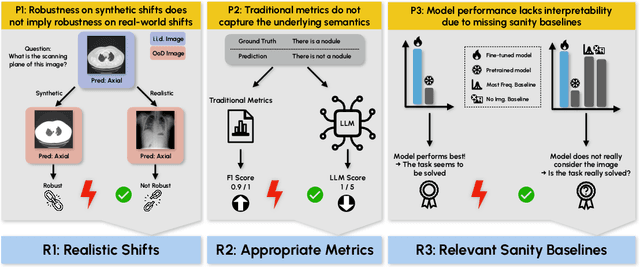
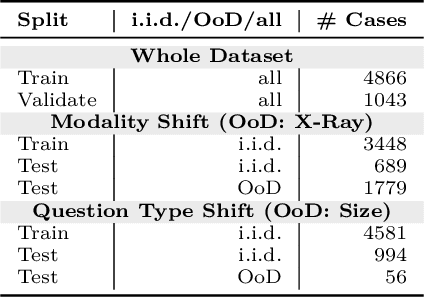
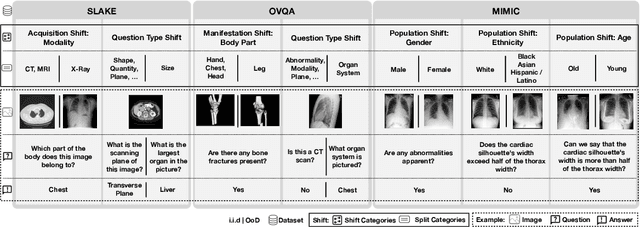
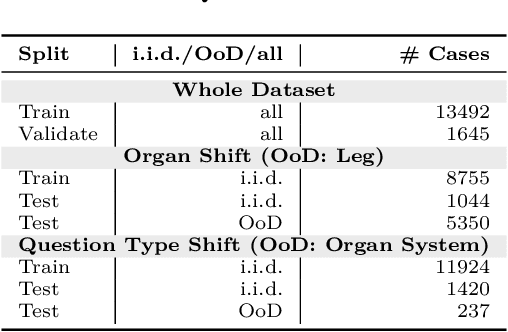
Abstract:Vision-Language Models (VLMs) have great potential in medical tasks, like Visual Question Answering (VQA), where they could act as interactive assistants for both patients and clinicians. Yet their robustness to distribution shifts on unseen data remains a critical concern for safe deployment. Evaluating such robustness requires a controlled experimental setup that allows for systematic insights into the model's behavior. However, we demonstrate that current setups fail to offer sufficiently thorough evaluations, limiting their ability to accurately assess model robustness. To address this gap, our work introduces a novel framework, called SURE-VQA, centered around three key requirements to overcome the current pitfalls and systematically analyze the robustness of VLMs: 1) Since robustness on synthetic shifts does not necessarily translate to real-world shifts, robustness should be measured on real-world shifts that are inherent to the VQA data; 2) Traditional token-matching metrics often fail to capture underlying semantics, necessitating the use of large language models (LLMs) for more accurate semantic evaluation; 3) Model performance often lacks interpretability due to missing sanity baselines, thus meaningful baselines should be reported that allow assessing the multimodal impact on the VLM. To demonstrate the relevance of this framework, we conduct a study on the robustness of various fine-tuning methods across three medical datasets with four different types of distribution shifts. Our study reveals several important findings: 1) Sanity baselines that do not utilize image data can perform surprisingly well; 2) We confirm LoRA as the best-performing PEFT method; 3) No PEFT method consistently outperforms others in terms of robustness to shifts. Code is provided at https://github.com/IML-DKFZ/sure-vqa.
Confidence intervals uncovered: Are we ready for real-world medical imaging AI?
Sep 27, 2024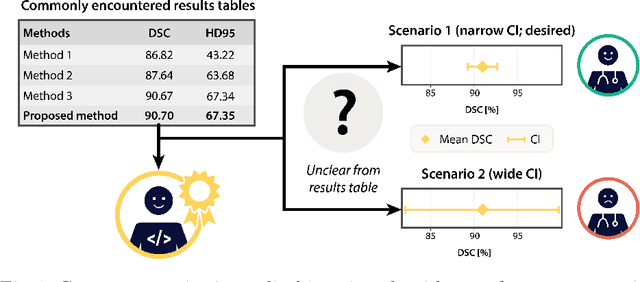
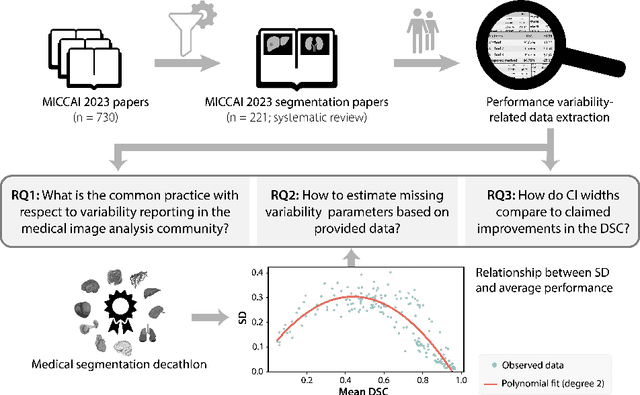
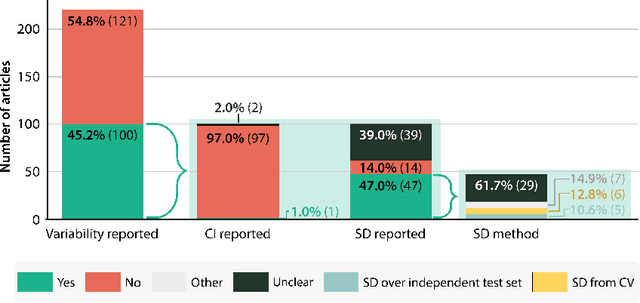

Abstract:Medical imaging is spearheading the AI transformation of healthcare. Performance reporting is key to determine which methods should be translated into clinical practice. Frequently, broad conclusions are simply derived from mean performance values. In this paper, we argue that this common practice is often a misleading simplification as it ignores performance variability. Our contribution is threefold. (1) Analyzing all MICCAI segmentation papers (n = 221) published in 2023, we first observe that more than 50% of papers do not assess performance variability at all. Moreover, only one (0.5%) paper reported confidence intervals (CIs) for model performance. (2) To address the reporting bottleneck, we show that the unreported standard deviation (SD) in segmentation papers can be approximated by a second-order polynomial function of the mean Dice similarity coefficient (DSC). Based on external validation data from 56 previous MICCAI challenges, we demonstrate that this approximation can accurately reconstruct the CI of a method using information provided in publications. (3) Finally, we reconstructed 95% CIs around the mean DSC of MICCAI 2023 segmentation papers. The median CI width was 0.03 which is three times larger than the median performance gap between the first and second ranked method. For more than 60% of papers, the mean performance of the second-ranked method was within the CI of the first-ranked method. We conclude that current publications typically do not provide sufficient evidence to support which models could potentially be translated into clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge