Lars Krämer
Finally Outshining the Random Baseline: A Simple and Effective Solution for Active Learning in 3D Biomedical Imaging
Jan 20, 2026Abstract:Active learning (AL) has the potential to drastically reduce annotation costs in 3D biomedical image segmentation, where expert labeling of volumetric data is both time-consuming and expensive. Yet, existing AL methods are unable to consistently outperform improved random sampling baselines adapted to 3D data, leaving the field without a reliable solution. We introduce Class-stratified Scheduled Power Predictive Entropy (ClaSP PE), a simple and effective query strategy that addresses two key limitations of standard uncertainty-based AL methods: class imbalance and redundancy in early selections. ClaSP PE combines class-stratified querying to ensure coverage of underrepresented structures and log-scale power noising with a decaying schedule to enforce query diversity in early-stage AL and encourage exploitation later. In our evaluation on 24 experimental settings using four 3D biomedical datasets within the comprehensive nnActive benchmark, ClaSP PE is the only method that generally outperforms improved random baselines in terms of both segmentation quality with statistically significant gains, whilst remaining annotation efficient. Furthermore, we explicitly simulate the real-world application by testing our method on four previously unseen datasets without manual adaptation, where all experiment parameters are set according to predefined guidelines. The results confirm that ClaSP PE robustly generalizes to novel tasks without requiring dataset-specific tuning. Within the nnActive framework, we present compelling evidence that an AL method can consistently outperform random baselines adapted to 3D segmentation, in terms of both performance and annotation efficiency in a realistic, close-to-production scenario. Our open-source implementation and clear deployment guidelines make it readily applicable in practice. Code is at https://github.com/MIC-DKFZ/nnActive.
nnInteractive: Redefining 3D Promptable Segmentation
Mar 11, 2025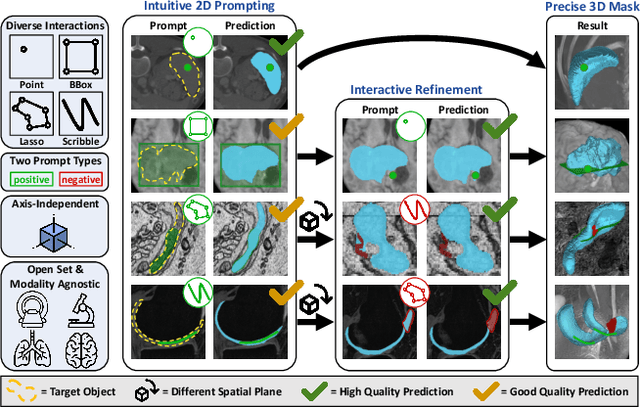
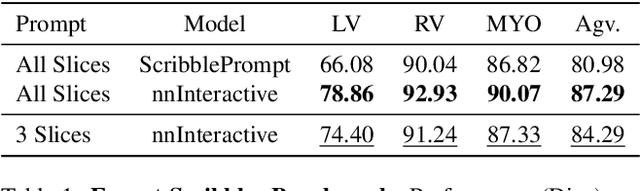
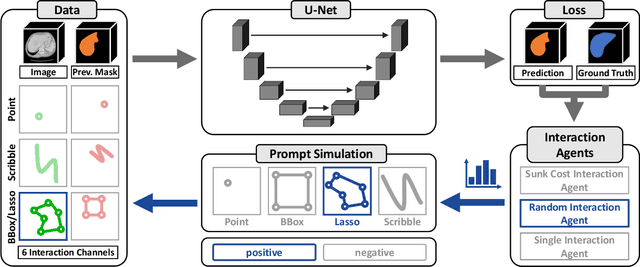
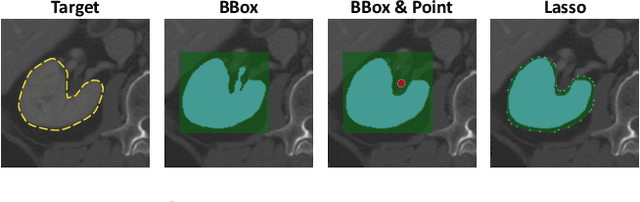
Abstract:Accurate and efficient 3D segmentation is essential for both clinical and research applications. While foundation models like SAM have revolutionized interactive segmentation, their 2D design and domain shift limitations make them ill-suited for 3D medical images. Current adaptations address some of these challenges but remain limited, either lacking volumetric awareness, offering restricted interactivity, or supporting only a small set of structures and modalities. Usability also remains a challenge, as current tools are rarely integrated into established imaging platforms and often rely on cumbersome web-based interfaces with restricted functionality. We introduce nnInteractive, the first comprehensive 3D interactive open-set segmentation method. It supports diverse prompts-including points, scribbles, boxes, and a novel lasso prompt-while leveraging intuitive 2D interactions to generate full 3D segmentations. Trained on 120+ diverse volumetric 3D datasets (CT, MRI, PET, 3D Microscopy, etc.), nnInteractive sets a new state-of-the-art in accuracy, adaptability, and usability. Crucially, it is the first method integrated into widely used image viewers (e.g., Napari, MITK), ensuring broad accessibility for real-world clinical and research applications. Extensive benchmarking demonstrates that nnInteractive far surpasses existing methods, setting a new standard for AI-driven interactive 3D segmentation. nnInteractive is publicly available: https://github.com/MIC-DKFZ/napari-nninteractive (Napari plugin), https://www.mitk.org/MITK-nnInteractive (MITK integration), https://github.com/MIC-DKFZ/nnInteractive (Python backend).
Code and Pixels: Multi-Modal Contrastive Pre-training for Enhanced Tabular Data Analysis
Jan 13, 2025



Abstract:Learning from tabular data is of paramount importance, as it complements the conventional analysis of image and video data by providing a rich source of structured information that is often critical for comprehensive understanding and decision-making processes. We present Multi-task Contrastive Masked Tabular Modeling (MT-CMTM), a novel method aiming to enhance tabular models by leveraging the correlation between tabular data and corresponding images. MT-CMTM employs a dual strategy combining contrastive learning with masked tabular modeling, optimizing the synergy between these data modalities. Central to our approach is a 1D Convolutional Neural Network with residual connections and an attention mechanism (1D-ResNet-CBAM), designed to efficiently process tabular data without relying on images. This enables MT-CMTM to handle purely tabular data for downstream tasks, eliminating the need for potentially costly image acquisition and processing. We evaluated MT-CMTM on the DVM car dataset, which is uniquely suited for this particular scenario, and the newly developed HIPMP dataset, which connects membrane fabrication parameters with image data. Our MT-CMTM model outperforms the proposed tabular 1D-ResNet-CBAM, which is trained from scratch, achieving a relative 1.48% improvement in relative MSE on HIPMP and a 2.38% increase in absolute accuracy on DVM. These results demonstrate MT-CMTM's robustness and its potential to advance the field of multi-modal learning.
Embarrassingly Simple Scribble Supervision for 3D Medical Segmentation
Mar 19, 2024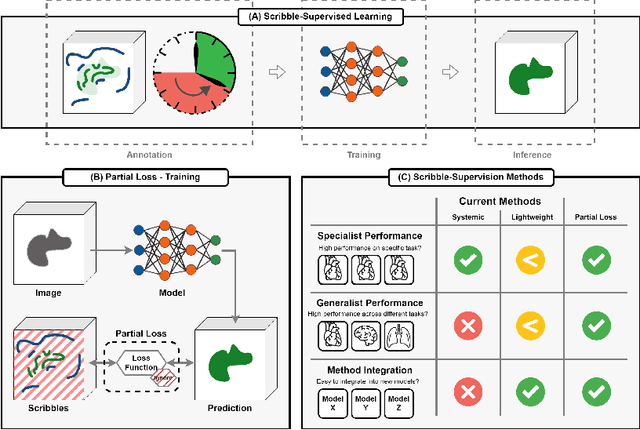
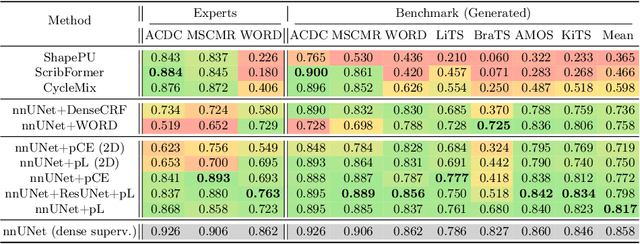
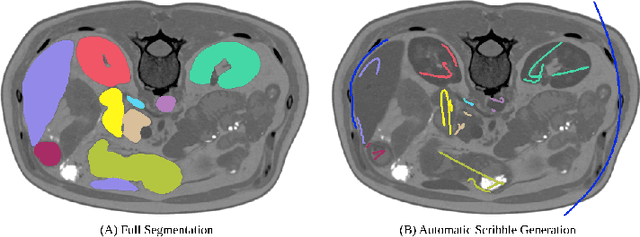
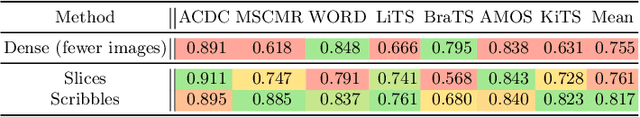
Abstract:Traditionally, segmentation algorithms require dense annotations for training, demanding significant annotation efforts, particularly within the 3D medical imaging field. Scribble-supervised learning emerges as a possible solution to this challenge, promising a reduction in annotation efforts when creating large-scale datasets. Recently, a plethora of methods for optimized learning from scribbles have been proposed, but have so far failed to position scribble annotation as a beneficial alternative. We relate this shortcoming to two major issues: 1) the complex nature of many methods which deeply ties them to the underlying segmentation model, thus preventing a migration to more powerful state-of-the-art models as the field progresses and 2) the lack of a systematic evaluation to validate consistent performance across the broader medical domain, resulting in a lack of trust when applying these methods to new segmentation problems. To address these issues, we propose a comprehensive scribble supervision benchmark consisting of seven datasets covering a diverse set of anatomies and pathologies imaged with varying modalities. We furthermore propose the systematic use of partial losses, i.e. losses that are only computed on annotated voxels. Contrary to most existing methods, these losses can be seamlessly integrated into state-of-the-art segmentation methods, enabling them to learn from scribble annotations while preserving their original loss formulations. Our evaluation using nnU-Net reveals that while most existing methods suffer from a lack of generalization, the proposed approach consistently delivers state-of-the-art performance. Thanks to its simplicity, our approach presents an embarrassingly simple yet effective solution to the challenges of scribble supervision. Source code as well as our extensive scribble benchmarking suite will be made publicly available upon publication.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge