Shuhan Xiao
nnLandmark: A Self-Configuring Method for 3D Medical Landmark Detection
Apr 10, 2025Abstract:Landmark detection plays a crucial role in medical imaging tasks that rely on precise spatial localization, including specific applications in diagnosis, treatment planning, image registration, and surgical navigation. However, manual annotation is labor-intensive and requires expert knowledge. While deep learning shows promise in automating this task, progress is hindered by limited public datasets, inconsistent benchmarks, and non-standardized baselines, restricting reproducibility, fair comparisons, and model generalizability. This work introduces nnLandmark, a self-configuring deep learning framework for 3D medical landmark detection, adapting nnU-Net to perform heatmap-based regression. By leveraging nnU-Net's automated configuration, nnLandmark eliminates the need for manual parameter tuning, offering out-of-the-box usability. It achieves state-of-the-art accuracy across two public datasets, with a mean radial error (MRE) of 1.5 mm on the Mandibular Molar Landmark (MML) dental CT dataset and 1.2 mm for anatomical fiducials on a brain MRI dataset (AFIDs), where nnLandmark aligns with the inter-rater variability of 1.5 mm. With its strong generalization, reproducibility, and ease of deployment, nnLandmark establishes a reliable baseline for 3D landmark detection, supporting research in anatomical localization and clinical workflows that depend on precise landmark identification. The code will be available soon.
Enhanced Diagnostic Fidelity in Pathology Whole Slide Image Compression via Deep Learning
Mar 14, 2025Abstract:Accurate diagnosis of disease often depends on the exhaustive examination of Whole Slide Images (WSI) at microscopic resolution. Efficient handling of these data-intensive images requires lossy compression techniques. This paper investigates the limitations of the widely-used JPEG algorithm, the current clinical standard, and reveals severe image artifacts impacting diagnostic fidelity. To overcome these challenges, we introduce a novel deep-learning (DL)-based compression method tailored for pathology images. By enforcing feature similarity of deep features between the original and compressed images, our approach achieves superior Peak Signal-to-Noise Ratio (PSNR), Multi-Scale Structural Similarity Index (MS-SSIM), and Learned Perceptual Image Patch Similarity (LPIPS) scores compared to JPEG-XL, Webp, and other DL compression methods.
Unlocking the Potential of Digital Pathology: Novel Baselines for Compression
Dec 17, 2024



Abstract:Digital pathology offers a groundbreaking opportunity to transform clinical practice in histopathological image analysis, yet faces a significant hurdle: the substantial file sizes of pathological Whole Slide Images (WSI). While current digital pathology solutions rely on lossy JPEG compression to address this issue, lossy compression can introduce color and texture disparities, potentially impacting clinical decision-making. While prior research addresses perceptual image quality and downstream performance independently of each other, we jointly evaluate compression schemes for perceptual and downstream task quality on four different datasets. In addition, we collect an initially uncompressed dataset for an unbiased perceptual evaluation of compression schemes. Our results show that deep learning models fine-tuned for perceptual quality outperform conventional compression schemes like JPEG-XL or WebP for further compression of WSI. However, they exhibit a significant bias towards the compression artifacts present in the training data and struggle to generalize across various compression schemes. We introduce a novel evaluation metric based on feature similarity between original files and compressed files that aligns very well with the actual downstream performance on the compressed WSI. Our metric allows for a general and standardized evaluation of lossy compression schemes and mitigates the requirement to independently assess different downstream tasks. Our study provides novel insights for the assessment of lossy compression schemes for WSI and encourages a unified evaluation of lossy compression schemes to accelerate the clinical uptake of digital pathology.
Data-Centric Strategies for Overcoming PET/CT Heterogeneity: Insights from the AutoPET III Lesion Segmentation Challenge
Sep 16, 2024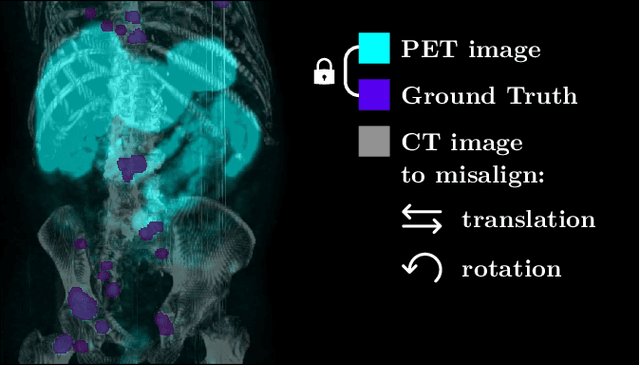

Abstract:The third autoPET challenge introduced a new data-centric task this year, shifting the focus from model development to improving metastatic lesion segmentation on PET/CT images through data quality and handling strategies. In response, we developed targeted methods to enhance segmentation performance tailored to the characteristics of PET/CT imaging. Our approach encompasses two key elements. First, to address potential alignment errors between CT and PET modalities as well as the prevalence of punctate lesions, we modified the baseline data augmentation scheme and extended it with misalignment augmentation. This adaptation aims to improve segmentation accuracy, particularly for tiny metastatic lesions. Second, to tackle the variability in image dimensions significantly affecting the prediction time, we implemented a dynamic ensembling and test-time augmentation (TTA) strategy. This method optimizes the use of ensembling and TTA within a 5-minute prediction time limit, effectively leveraging the generalization potential for both small and large images. Both of our solutions are designed to be robust across different tracers and institutional settings, offering a general, yet imaging-specific approach to the multi-tracer and multi-institutional challenges of the competition. We made the challenge repository with our modifications publicly available at \url{https://github.com/MIC-DKFZ/miccai2024_autopet3_datacentric}.
From FDG to PSMA: A Hitchhiker's Guide to Multitracer, Multicenter Lesion Segmentation in PET/CT Imaging
Sep 14, 2024

Abstract:Automated lesion segmentation in PET/CT scans is crucial for improving clinical workflows and advancing cancer diagnostics. However, the task is challenging due to physiological variability, different tracers used in PET imaging, and diverse imaging protocols across medical centers. To address this, the autoPET series was created to challenge researchers to develop algorithms that generalize across diverse PET/CT environments. This paper presents our solution for the autoPET III challenge, targeting multitracer, multicenter generalization using the nnU-Net framework with the ResEncL architecture. Key techniques include misalignment data augmentation and multi-modal pretraining across CT, MR, and PET datasets to provide an initial anatomical understanding. We incorporate organ supervision as a multitask approach, enabling the model to distinguish between physiological uptake and tracer-specific patterns, which is particularly beneficial in cases where no lesions are present. Compared to the default nnU-Net, which achieved a Dice score of 57.61, or the larger ResEncL (65.31) our model significantly improved performance with a Dice score of 68.40, alongside a reduction in false positive (FPvol: 7.82) and false negative (FNvol: 10.35) volumes. These results underscore the effectiveness of combining advanced network design, augmentation, pretraining, and multitask learning for PET/CT lesion segmentation. Code is publicly available at https://github.com/MIC-DKFZ/autopet-3-submission.
Learned Image Compression for HE-stained Histopathological Images via Stain Deconvolution
Jun 18, 2024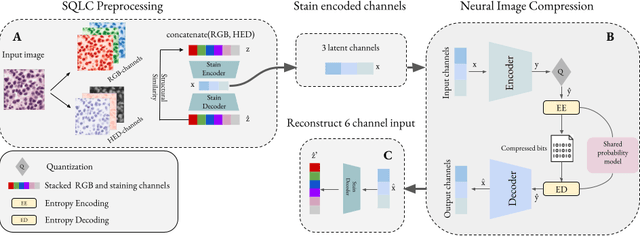
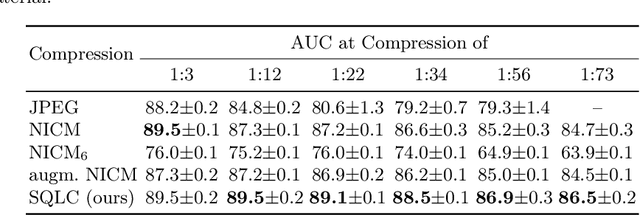
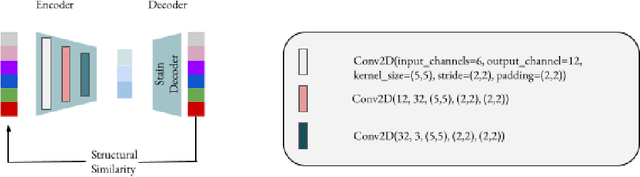
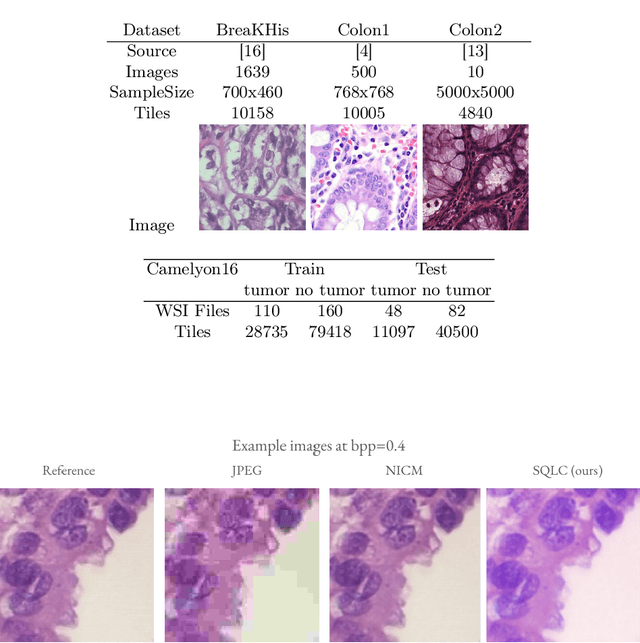
Abstract:Processing histopathological Whole Slide Images (WSI) leads to massive storage requirements for clinics worldwide. Even after lossy image compression during image acquisition, additional lossy compression is frequently possible without substantially affecting the performance of deep learning-based (DL) downstream tasks. In this paper, we show that the commonly used JPEG algorithm is not best suited for further compression and we propose Stain Quantized Latent Compression (SQLC ), a novel DL based histopathology data compression approach. SQLC compresses staining and RGB channels before passing it through a compression autoencoder (CAE ) in order to obtain quantized latent representations for maximizing the compression. We show that our approach yields superior performance in a classification downstream task, compared to traditional approaches like JPEG, while image quality metrics like the Multi-Scale Structural Similarity Index (MS-SSIM) is largely preserved. Our method is online available.
Enhancing predictive imaging biomarker discovery through treatment effect analysis
Jun 04, 2024Abstract:Identifying predictive biomarkers, which forecast individual treatment effectiveness, is crucial for personalized medicine and informs decision-making across diverse disciplines. These biomarkers are extracted from pre-treatment data, often within randomized controlled trials, and have to be distinguished from prognostic biomarkers, which are independent of treatment assignment. Our study focuses on the discovery of predictive imaging biomarkers, aiming to leverage pre-treatment images to unveil new causal relationships. Previous approaches relied on labor-intensive handcrafted or manually derived features, which may introduce biases. In response, we present a new task of discovering predictive imaging biomarkers directly from the pre-treatment images to learn relevant image features. We propose an evaluation protocol for this task to assess a model's ability to identify predictive imaging biomarkers and differentiate them from prognostic ones. It employs statistical testing and a comprehensive analysis of image feature attribution. We explore the suitability of deep learning models originally designed for estimating the conditional average treatment effect (CATE) for this task, which previously have been primarily assessed for the precision of CATE estimation, overlooking the evaluation of imaging biomarker discovery. Our proof-of-concept analysis demonstrates promising results in discovering and validating predictive imaging biomarkers from synthetic outcomes and real-world image datasets.
Embarrassingly Simple Scribble Supervision for 3D Medical Segmentation
Mar 19, 2024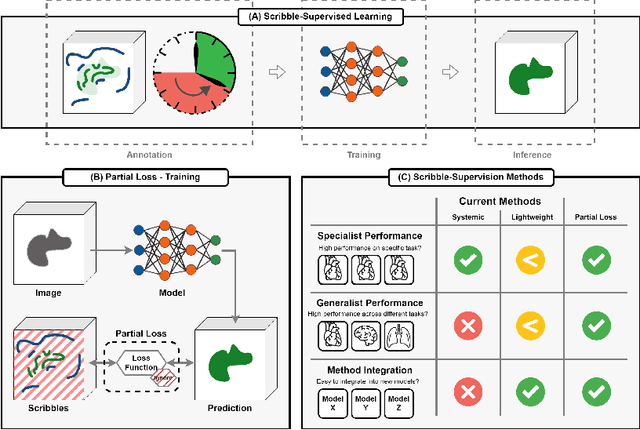
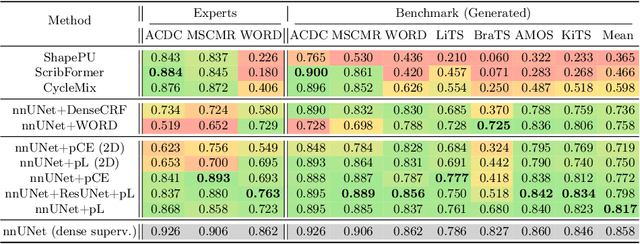
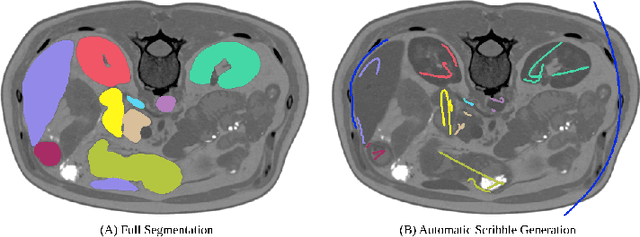
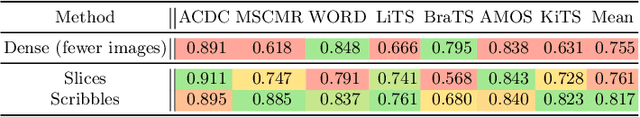
Abstract:Traditionally, segmentation algorithms require dense annotations for training, demanding significant annotation efforts, particularly within the 3D medical imaging field. Scribble-supervised learning emerges as a possible solution to this challenge, promising a reduction in annotation efforts when creating large-scale datasets. Recently, a plethora of methods for optimized learning from scribbles have been proposed, but have so far failed to position scribble annotation as a beneficial alternative. We relate this shortcoming to two major issues: 1) the complex nature of many methods which deeply ties them to the underlying segmentation model, thus preventing a migration to more powerful state-of-the-art models as the field progresses and 2) the lack of a systematic evaluation to validate consistent performance across the broader medical domain, resulting in a lack of trust when applying these methods to new segmentation problems. To address these issues, we propose a comprehensive scribble supervision benchmark consisting of seven datasets covering a diverse set of anatomies and pathologies imaged with varying modalities. We furthermore propose the systematic use of partial losses, i.e. losses that are only computed on annotated voxels. Contrary to most existing methods, these losses can be seamlessly integrated into state-of-the-art segmentation methods, enabling them to learn from scribble annotations while preserving their original loss formulations. Our evaluation using nnU-Net reveals that while most existing methods suffer from a lack of generalization, the proposed approach consistently delivers state-of-the-art performance. Thanks to its simplicity, our approach presents an embarrassingly simple yet effective solution to the challenges of scribble supervision. Source code as well as our extensive scribble benchmarking suite will be made publicly available upon publication.
Leveraging Foundation Models for Content-Based Medical Image Retrieval in Radiology
Mar 11, 2024Abstract:Content-based image retrieval (CBIR) has the potential to significantly improve diagnostic aid and medical research in radiology. Current CBIR systems face limitations due to their specialization to certain pathologies, limiting their utility. In response, we propose using vision foundation models as powerful and versatile off-the-shelf feature extractors for content-based medical image retrieval. By benchmarking these models on a comprehensive dataset of 1.6 million 2D radiological images spanning four modalities and 161 pathologies, we identify weakly-supervised models as superior, achieving a P@1 of up to 0.594. This performance not only competes with a specialized model but does so without the need for fine-tuning. Our analysis further explores the challenges in retrieving pathological versus anatomical structures, indicating that accurate retrieval of pathological features presents greater difficulty. Despite these challenges, our research underscores the vast potential of foundation models for CBIR in radiology, proposing a shift towards versatile, general-purpose medical image retrieval systems that do not require specific tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge