Peter Schüffler
Dataset Distillation with Probabilistic Latent Features
May 10, 2025Abstract:As deep learning models grow in complexity and the volume of training data increases, reducing storage and computational costs becomes increasingly important. Dataset distillation addresses this challenge by synthesizing a compact set of synthetic data that can effectively replace the original dataset in downstream classification tasks. While existing methods typically rely on mapping data from pixel space to the latent space of a generative model, we propose a novel stochastic approach that models the joint distribution of latent features. This allows our method to better capture spatial structures and produce diverse synthetic samples, which benefits model training. Specifically, we introduce a low-rank multivariate normal distribution parameterized by a lightweight network. This design maintains low computational complexity and is compatible with various matching networks used in dataset distillation. After distillation, synthetic images are generated by feeding the learned latent features into a pretrained generator. These synthetic images are then used to train classification models, and performance is evaluated on real test set. We validate our method on several benchmarks, including ImageNet subsets, CIFAR-10, and the MedMNIST histopathological dataset. Our approach achieves state-of-the-art cross architecture performance across a range of backbone architectures, demonstrating its generality and effectiveness.
From Pixels to Histopathology: A Graph-Based Framework for Interpretable Whole Slide Image Analysis
Mar 14, 2025Abstract:The histopathological classification of whole-slide images (WSIs) is a fundamental task in digital pathology; yet it requires extensive time and expertise from specialists. While deep learning methods show promising results, they typically process WSIs by dividing them into artificial patches, which inherently prevents a network from learning from the entire image context, disregards natural tissue structures and compromises interpretability. Our method overcomes this limitation through a novel graph-based framework that constructs WSI graph representations. The WSI-graph efficiently captures essential histopathological information in a compact form. We build tissue representations (nodes) that follow biological boundaries rather than arbitrary patches all while providing interpretable features for explainability. Through adaptive graph coarsening guided by learned embeddings, we progressively merge regions while maintaining discriminative local features and enabling efficient global information exchange. In our method's final step, we solve the diagnostic task through a graph attention network. We empirically demonstrate strong performance on multiple challenging tasks such as cancer stage classification and survival prediction, while also identifying predictive factors using Integrated Gradients. Our implementation is publicly available at https://github.com/HistoGraph31/pix2pathology
Enhanced Diagnostic Fidelity in Pathology Whole Slide Image Compression via Deep Learning
Mar 14, 2025Abstract:Accurate diagnosis of disease often depends on the exhaustive examination of Whole Slide Images (WSI) at microscopic resolution. Efficient handling of these data-intensive images requires lossy compression techniques. This paper investigates the limitations of the widely-used JPEG algorithm, the current clinical standard, and reveals severe image artifacts impacting diagnostic fidelity. To overcome these challenges, we introduce a novel deep-learning (DL)-based compression method tailored for pathology images. By enforcing feature similarity of deep features between the original and compressed images, our approach achieves superior Peak Signal-to-Noise Ratio (PSNR), Multi-Scale Structural Similarity Index (MS-SSIM), and Learned Perceptual Image Patch Similarity (LPIPS) scores compared to JPEG-XL, Webp, and other DL compression methods.
Unlocking the Potential of Digital Pathology: Novel Baselines for Compression
Dec 17, 2024



Abstract:Digital pathology offers a groundbreaking opportunity to transform clinical practice in histopathological image analysis, yet faces a significant hurdle: the substantial file sizes of pathological Whole Slide Images (WSI). While current digital pathology solutions rely on lossy JPEG compression to address this issue, lossy compression can introduce color and texture disparities, potentially impacting clinical decision-making. While prior research addresses perceptual image quality and downstream performance independently of each other, we jointly evaluate compression schemes for perceptual and downstream task quality on four different datasets. In addition, we collect an initially uncompressed dataset for an unbiased perceptual evaluation of compression schemes. Our results show that deep learning models fine-tuned for perceptual quality outperform conventional compression schemes like JPEG-XL or WebP for further compression of WSI. However, they exhibit a significant bias towards the compression artifacts present in the training data and struggle to generalize across various compression schemes. We introduce a novel evaluation metric based on feature similarity between original files and compressed files that aligns very well with the actual downstream performance on the compressed WSI. Our metric allows for a general and standardized evaluation of lossy compression schemes and mitigates the requirement to independently assess different downstream tasks. Our study provides novel insights for the assessment of lossy compression schemes for WSI and encourages a unified evaluation of lossy compression schemes to accelerate the clinical uptake of digital pathology.
Learned Image Compression for HE-stained Histopathological Images via Stain Deconvolution
Jun 18, 2024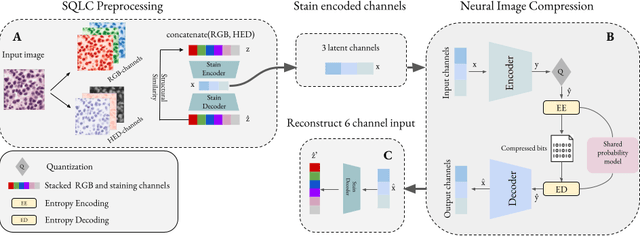
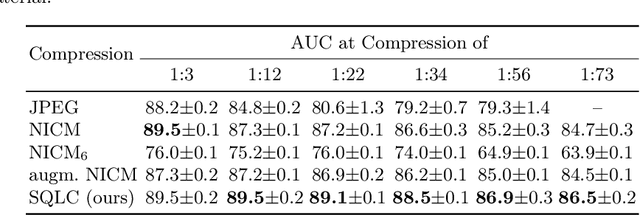
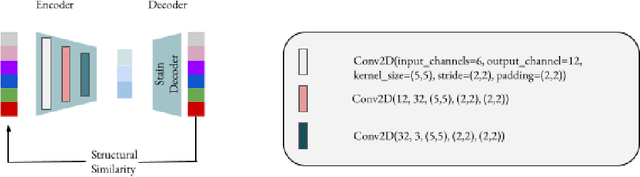
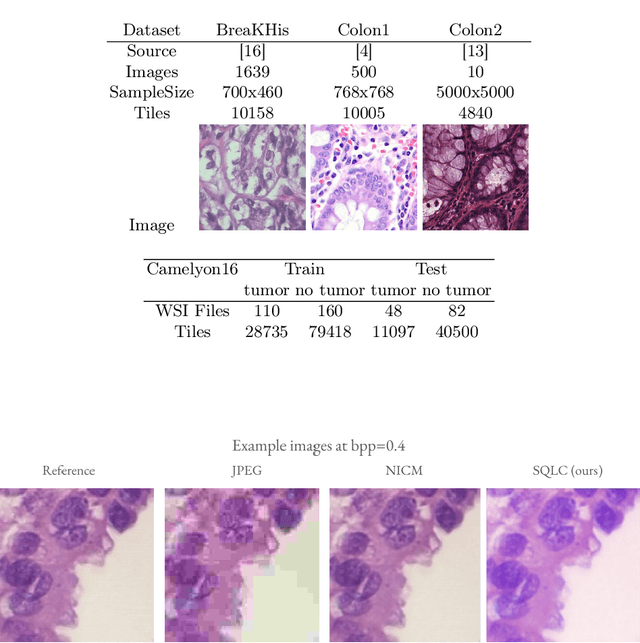
Abstract:Processing histopathological Whole Slide Images (WSI) leads to massive storage requirements for clinics worldwide. Even after lossy image compression during image acquisition, additional lossy compression is frequently possible without substantially affecting the performance of deep learning-based (DL) downstream tasks. In this paper, we show that the commonly used JPEG algorithm is not best suited for further compression and we propose Stain Quantized Latent Compression (SQLC ), a novel DL based histopathology data compression approach. SQLC compresses staining and RGB channels before passing it through a compression autoencoder (CAE ) in order to obtain quantized latent representations for maximizing the compression. We show that our approach yields superior performance in a classification downstream task, compared to traditional approaches like JPEG, while image quality metrics like the Multi-Scale Structural Similarity Index (MS-SSIM) is largely preserved. Our method is online available.
Whole Slide Multiple Instance Learning for Predicting Axillary Lymph Node Metastasis
Oct 06, 2023Abstract:Breast cancer is a major concern for women's health globally, with axillary lymph node (ALN) metastasis identification being critical for prognosis evaluation and treatment guidance. This paper presents a deep learning (DL) classification pipeline for quantifying clinical information from digital core-needle biopsy (CNB) images, with one step less than existing methods. A publicly available dataset of 1058 patients was used to evaluate the performance of different baseline state-of-the-art (SOTA) DL models in classifying ALN metastatic status based on CNB images. An extensive ablation study of various data augmentation techniques was also conducted. Finally, the manual tumor segmentation and annotation step performed by the pathologists was assessed.
* Accepted for MICCAI DEMI Workshop 2023
DISBELIEVE: Distance Between Client Models is Very Essential for Effective Local Model Poisoning Attacks
Aug 14, 2023Abstract:Federated learning is a promising direction to tackle the privacy issues related to sharing patients' sensitive data. Often, federated systems in the medical image analysis domain assume that the participating local clients are \textit{honest}. Several studies report mechanisms through which a set of malicious clients can be introduced that can poison the federated setup, hampering the performance of the global model. To overcome this, robust aggregation methods have been proposed that defend against those attacks. We observe that most of the state-of-the-art robust aggregation methods are heavily dependent on the distance between the parameters or gradients of malicious clients and benign clients, which makes them prone to local model poisoning attacks when the parameters or gradients of malicious and benign clients are close. Leveraging this, we introduce DISBELIEVE, a local model poisoning attack that creates malicious parameters or gradients such that their distance to benign clients' parameters or gradients is low respectively but at the same time their adverse effect on the global model's performance is high. Experiments on three publicly available medical image datasets demonstrate the efficacy of the proposed DISBELIEVE attack as it significantly lowers the performance of the state-of-the-art \textit{robust aggregation} methods for medical image analysis. Furthermore, compared to state-of-the-art local model poisoning attacks, DISBELIEVE attack is also effective on natural images where we observe a severe drop in classification performance of the global model for multi-class classification on benchmark dataset CIFAR-10.
Multi-Organ Cancer Classification and Survival Analysis
Dec 02, 2016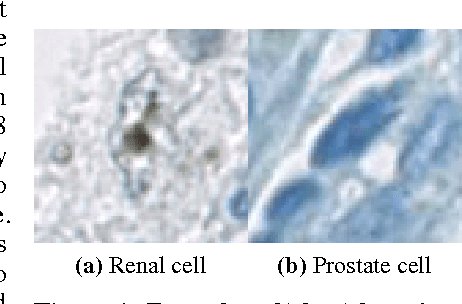



Abstract:Accurate and robust cell nuclei classification is the cornerstone for a wider range of tasks in digital and Computational Pathology. However, most machine learning systems require extensive labeling from expert pathologists for each individual problem at hand, with no or limited abilities for knowledge transfer between datasets and organ sites. In this paper we implement and evaluate a variety of deep neural network models and model ensembles for nuclei classification in renal cell cancer (RCC) and prostate cancer (PCa). We propose a convolutional neural network system based on residual learning which significantly improves over the state-of-the-art in cell nuclei classification. Finally, we show that the combination of tissue types during training increases not only classification accuracy but also overall survival analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge