Peter Neher
Kaapana: A Comprehensive Open-Source Platform for Integrating AI in Medical Imaging Research Environments
Dec 10, 2025
Abstract:Developing generalizable AI for medical imaging requires both access to large, multi-center datasets and standardized, reproducible tooling within research environments. However, leveraging real-world imaging data in clinical research environments is still hampered by strict regulatory constraints, fragmented software infrastructure, and the challenges inherent in conducting large-cohort multicentre studies. This leads to projects that rely on ad-hoc toolchains that are hard to reproduce, difficult to scale beyond single institutions and poorly suited for collaboration between clinicians and data scientists. We present Kaapana, a comprehensive open-source platform for medical imaging research that is designed to bridge this gap. Rather than building single-use, site-specific tooling, Kaapana provides a modular, extensible framework that unifies data ingestion, cohort curation, processing workflows and result inspection under a common user interface. By bringing the algorithm to the data, it enables institutions to keep control over their sensitive data while still participating in distributed experimentation and model development. By integrating flexible workflow orchestration with user-facing applications for researchers, Kaapana reduces technical overhead, improves reproducibility and enables conducting large-scale, collaborative, multi-centre imaging studies. We describe the core concepts of the platform and illustrate how they can support diverse use cases, from local prototyping to nation-wide research networks. The open-source codebase is available at https://github.com/kaapana/kaapana
nnLandmark: A Self-Configuring Method for 3D Medical Landmark Detection
Apr 10, 2025Abstract:Landmark detection plays a crucial role in medical imaging tasks that rely on precise spatial localization, including specific applications in diagnosis, treatment planning, image registration, and surgical navigation. However, manual annotation is labor-intensive and requires expert knowledge. While deep learning shows promise in automating this task, progress is hindered by limited public datasets, inconsistent benchmarks, and non-standardized baselines, restricting reproducibility, fair comparisons, and model generalizability. This work introduces nnLandmark, a self-configuring deep learning framework for 3D medical landmark detection, adapting nnU-Net to perform heatmap-based regression. By leveraging nnU-Net's automated configuration, nnLandmark eliminates the need for manual parameter tuning, offering out-of-the-box usability. It achieves state-of-the-art accuracy across two public datasets, with a mean radial error (MRE) of 1.5 mm on the Mandibular Molar Landmark (MML) dental CT dataset and 1.2 mm for anatomical fiducials on a brain MRI dataset (AFIDs), where nnLandmark aligns with the inter-rater variability of 1.5 mm. With its strong generalization, reproducibility, and ease of deployment, nnLandmark establishes a reliable baseline for 3D landmark detection, supporting research in anatomical localization and clinical workflows that depend on precise landmark identification. The code will be available soon.
Enhanced Diagnostic Fidelity in Pathology Whole Slide Image Compression via Deep Learning
Mar 14, 2025Abstract:Accurate diagnosis of disease often depends on the exhaustive examination of Whole Slide Images (WSI) at microscopic resolution. Efficient handling of these data-intensive images requires lossy compression techniques. This paper investigates the limitations of the widely-used JPEG algorithm, the current clinical standard, and reveals severe image artifacts impacting diagnostic fidelity. To overcome these challenges, we introduce a novel deep-learning (DL)-based compression method tailored for pathology images. By enforcing feature similarity of deep features between the original and compressed images, our approach achieves superior Peak Signal-to-Noise Ratio (PSNR), Multi-Scale Structural Similarity Index (MS-SSIM), and Learned Perceptual Image Patch Similarity (LPIPS) scores compared to JPEG-XL, Webp, and other DL compression methods.
Precision ICU Resource Planning: A Multimodal Model for Brain Surgery Outcomes
Dec 20, 2024

Abstract:Although advances in brain surgery techniques have led to fewer postoperative complications requiring Intensive Care Unit (ICU) monitoring, the routine transfer of patients to the ICU remains the clinical standard, despite its high cost. Predictive Gradient Boosted Trees based on clinical data have attempted to optimize ICU admission by identifying key risk factors pre-operatively; however, these approaches overlook valuable imaging data that could enhance prediction accuracy. In this work, we show that multimodal approaches that combine clinical data with imaging data outperform the current clinical data only baseline from 0.29 [F1] to 0.30 [F1], when only pre-operative clinical data is used and from 0.37 [F1] to 0.41 [F1], for pre- and post-operative data. This study demonstrates that effective ICU admission prediction benefits from multimodal data fusion, especially in contexts of severe class imbalance.
Unlocking the Potential of Digital Pathology: Novel Baselines for Compression
Dec 17, 2024



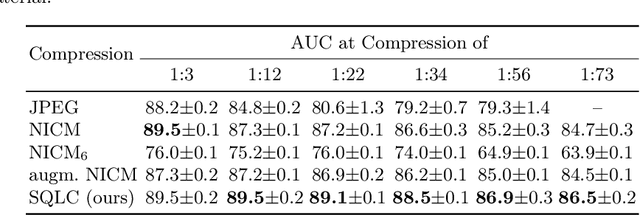
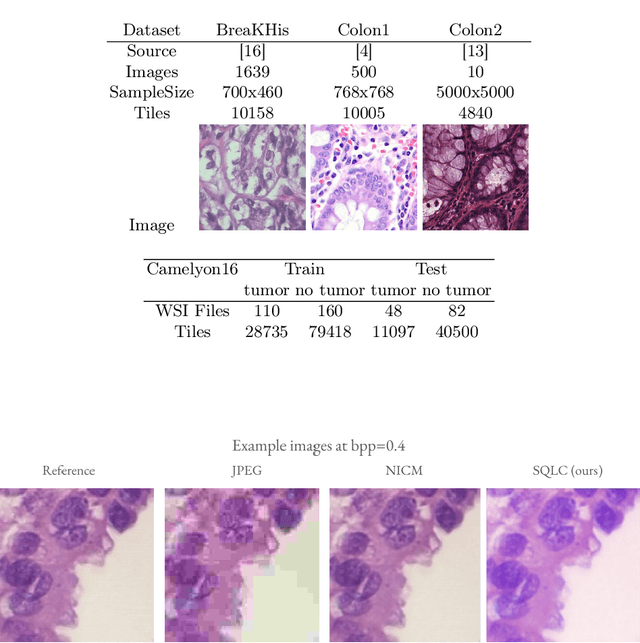
Abstract:Digital pathology offers a groundbreaking opportunity to transform clinical practice in histopathological image analysis, yet faces a significant hurdle: the substantial file sizes of pathological Whole Slide Images (WSI). While current digital pathology solutions rely on lossy JPEG compression to address this issue, lossy compression can introduce color and texture disparities, potentially impacting clinical decision-making. While prior research addresses perceptual image quality and downstream performance independently of each other, we jointly evaluate compression schemes for perceptual and downstream task quality on four different datasets. In addition, we collect an initially uncompressed dataset for an unbiased perceptual evaluation of compression schemes. Our results show that deep learning models fine-tuned for perceptual quality outperform conventional compression schemes like JPEG-XL or WebP for further compression of WSI. However, they exhibit a significant bias towards the compression artifacts present in the training data and struggle to generalize across various compression schemes. We introduce a novel evaluation metric based on feature similarity between original files and compressed files that aligns very well with the actual downstream performance on the compressed WSI. Our metric allows for a general and standardized evaluation of lossy compression schemes and mitigates the requirement to independently assess different downstream tasks. Our study provides novel insights for the assessment of lossy compression schemes for WSI and encourages a unified evaluation of lossy compression schemes to accelerate the clinical uptake of digital pathology.
Learned Image Compression for HE-stained Histopathological Images via Stain Deconvolution
Jun 18, 2024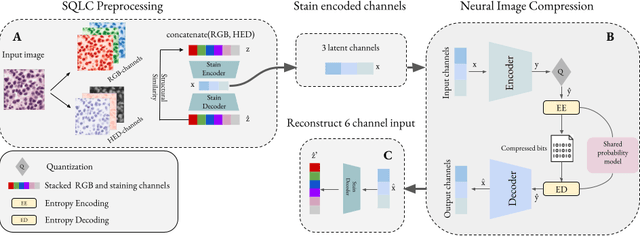
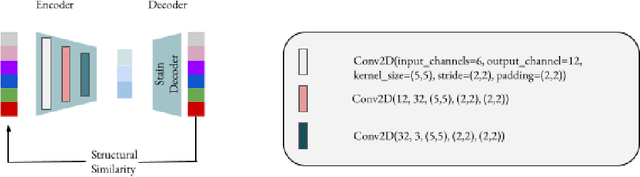
Abstract:Processing histopathological Whole Slide Images (WSI) leads to massive storage requirements for clinics worldwide. Even after lossy image compression during image acquisition, additional lossy compression is frequently possible without substantially affecting the performance of deep learning-based (DL) downstream tasks. In this paper, we show that the commonly used JPEG algorithm is not best suited for further compression and we propose Stain Quantized Latent Compression (SQLC ), a novel DL based histopathology data compression approach. SQLC compresses staining and RGB channels before passing it through a compression autoencoder (CAE ) in order to obtain quantized latent representations for maximizing the compression. We show that our approach yields superior performance in a classification downstream task, compared to traditional approaches like JPEG, while image quality metrics like the Multi-Scale Structural Similarity Index (MS-SSIM) is largely preserved. Our method is online available.
Real-World Federated Learning in Radiology: Hurdles to overcome and Benefits to gain
May 15, 2024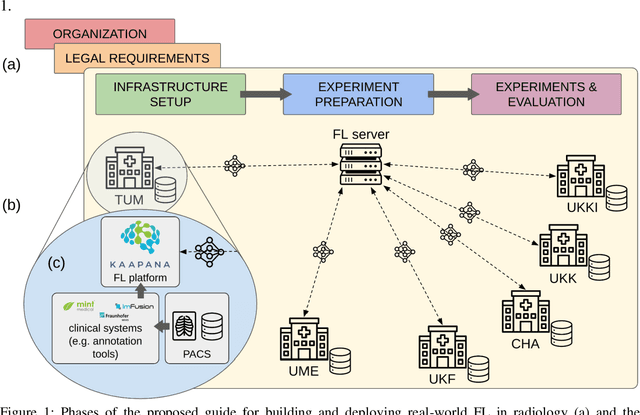



Abstract:Objective: Federated Learning (FL) enables collaborative model training while keeping data locally. Currently, most FL studies in radiology are conducted in simulated environments due to numerous hurdles impeding its translation into practice. The few existing real-world FL initiatives rarely communicate specific measures taken to overcome these hurdles, leaving behind a significant knowledge gap. Minding efforts to implement real-world FL, there is a notable lack of comprehensive assessment comparing FL to less complex alternatives. Materials & Methods: We extensively reviewed FL literature, categorizing insights along with our findings according to their nature and phase while establishing a FL initiative, summarized to a comprehensive guide. We developed our own FL infrastructure within the German Radiological Cooperative Network (RACOON) and demonstrated its functionality by training FL models on lung pathology segmentation tasks across six university hospitals. We extensively evaluated FL against less complex alternatives in three distinct evaluation scenarios. Results: The proposed guide outlines essential steps, identified hurdles, and proposed solutions for establishing successful FL initiatives conducting real-world experiments. Our experimental results show that FL outperforms less complex alternatives in all evaluation scenarios, justifying the effort required to translate FL into real-world applications. Discussion & Conclusion: Our proposed guide aims to aid future FL researchers in circumventing pitfalls and accelerating translation of FL into radiological applications. Our results underscore the value of efforts needed to translate FL into real-world applications by demonstrating advantageous performance over alternatives, and emphasize the importance of strategic organization, robust management of distributed data and infrastructure in real-world settings.
atTRACTive: Semi-automatic white matter tract segmentation using active learning
May 31, 2023



Abstract:Accurately identifying white matter tracts in medical images is essential for various applications, including surgery planning and tract-specific analysis. Supervised machine learning models have reached state-of-the-art solving this task automatically. However, these models are primarily trained on healthy subjects and struggle with strong anatomical aberrations, e.g. caused by brain tumors. This limitation makes them unsuitable for tasks such as preoperative planning, wherefore time-consuming and challenging manual delineation of the target tract is typically employed. We propose semi-automatic entropy-based active learning for quick and intuitive segmentation of white matter tracts from whole-brain tractography consisting of millions of streamlines. The method is evaluated on 21 openly available healthy subjects from the Human Connectome Project and an internal dataset of ten neurosurgical cases. With only a few annotations, the proposed approach enables segmenting tracts on tumor cases comparable to healthy subjects (dice=0.71), while the performance of automatic methods, like TractSeg dropped substantially (dice=0.34) in comparison to healthy subjects. The method is implemented as a prototype named atTRACTive in the freely available software MITK Diffusion. Manual experiments on tumor data showed higher efficiency due to lower segmentation times compared to traditional ROI-based segmentation.
Combined tract segmentation and orientation mapping for bundle-specific tractography
Jan 29, 2019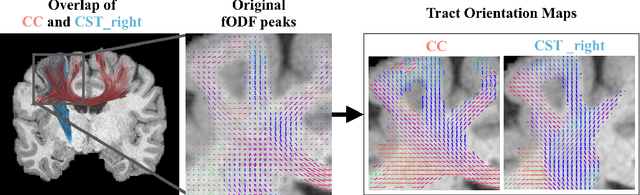
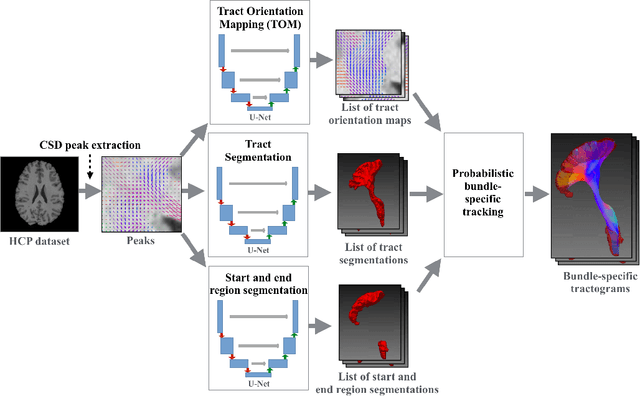
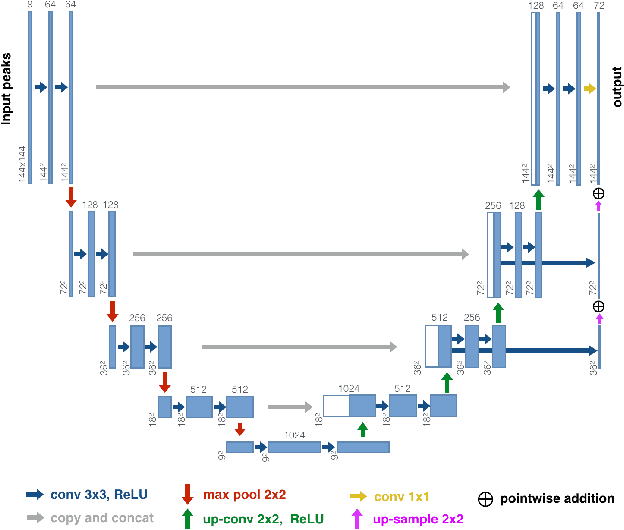
Abstract:While the major white matter tracts are of great interest to numerous studies in neuroscience and medicine, their manual dissection in larger cohorts from diffusion MRI tractograms is time-consuming, requires expert knowledge and is hard to reproduce. In previous work we presented tract orientation mapping (TOM) as a novel concept for bundle-specific tractography. It is based on a learned mapping from the original fiber orientation distribution function (fODF) peaks to tract specific peaks, called tract orientation maps. Each tract orientation map represents the voxel-wise principal orientation of one tract.Here, we present an extension of this approach that combines TOM with accurate segmentations of the tract outline and its start and end region. We also introduce a custom probabilistic tracking algorithm that samples from a Gaussian distribution with fixed standard deviation centered on each peak thus enabling more complete trackings on the tract orientation maps than deterministic tracking. These extensions enable the automatic creation of bundle-specific tractograms with previously unseen accuracy. We show for 72 different bundles on high quality, low quality and phantom data that our approach runs faster and produces more accurate bundle-specific tractograms than 7 state of the art benchmark methods while avoiding cumbersome processing steps like whole brain tractography, non-linear registration, clustering or manual dissection. Moreover, we show on 17 datasets that our approach generalizes well to datasets acquired with different scanners and settings as well as with pathologies. The code of our method is openly available at www.github.com/MIC-DKFZ/TractSeg.
TractSeg - Fast and accurate white matter tract segmentation
Aug 20, 2018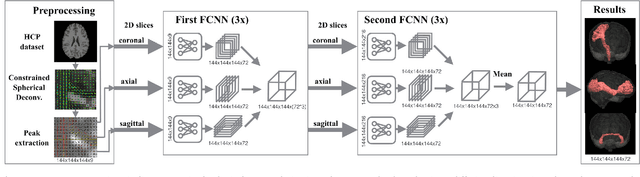
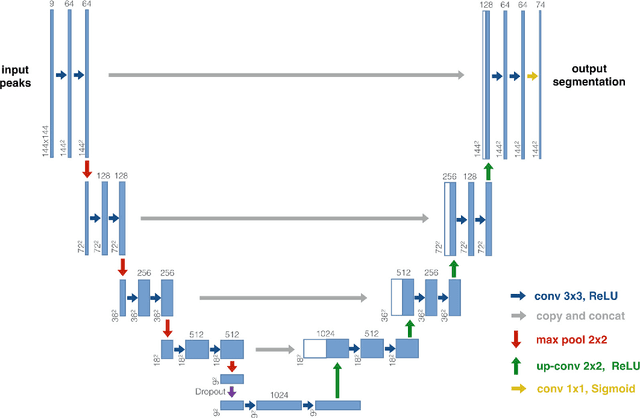
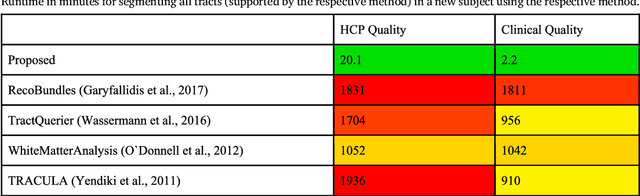
Abstract:The individual course of white matter fiber tracts is an important key for analysis of white matter characteristics in healthy and diseased brains. Uniquely, diffusion-weighted MRI tractography in combination with region-based or clustering-based selection of streamlines allows for the in-vivo delineation and analysis of anatomically well known tracts. This, however, currently requires complex, computationally intensive and tedious-to-set-up processing pipelines. TractSeg is a novel convolutional neural network-based approach that directly segments tracts in the field of fiber orientation distribution function (fODF) peaks without requiring tractography, image registration or parcellation. We demonstrate in 105 subjects from the Human Connectome Project that the proposed approach is much faster than existing methods while providing unprecedented accuracy. The code and data are openly available at https://github.com/MIC-DKFZ/TractSeg/ and https://doi.org/10.5281/zenodo.1088277, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge