Jakob Wasserthal
In search of truth: Evaluating concordance of AI-based anatomy segmentation models
Dec 17, 2025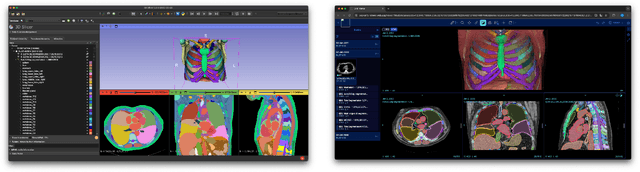


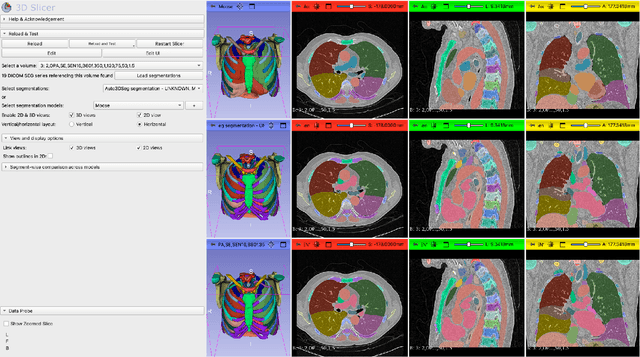
Abstract:Purpose AI-based methods for anatomy segmentation can help automate characterization of large imaging datasets. The growing number of similar in functionality models raises the challenge of evaluating them on datasets that do not contain ground truth annotations. We introduce a practical framework to assist in this task. Approach We harmonize the segmentation results into a standard, interoperable representation, which enables consistent, terminology-based labeling of the structures. We extend 3D Slicer to streamline loading and comparison of these harmonized segmentations, and demonstrate how standard representation simplifies review of the results using interactive summary plots and browser-based visualization using OHIF Viewer. To demonstrate the utility of the approach we apply it to evaluating segmentation of 31 anatomical structures (lungs, vertebrae, ribs, and heart) by six open-source models - TotalSegmentator 1.5 and 2.6, Auto3DSeg, MOOSE, MultiTalent, and CADS - for a sample of Computed Tomography (CT) scans from the publicly available National Lung Screening Trial (NLST) dataset. Results We demonstrate the utility of the framework in enabling automating loading, structure-wise inspection and comparison across models. Preliminary results ascertain practical utility of the approach in allowing quick detection and review of problematic results. The comparison shows excellent agreement segmenting some (e.g., lung) but not all structures (e.g., some models produce invalid vertebrae or rib segmentations). Conclusions The resources developed are linked from https://imagingdatacommons.github.io/segmentation-comparison/ including segmentation harmonization scripts, summary plots, and visualization tools. This work assists in model evaluation in absence of ground truth, ultimately enabling informed model selection.
Multi-centric AI Model for Unruptured Intracranial Aneurysm Detection and Volumetric Segmentation in 3D TOF-MRI
Aug 30, 2024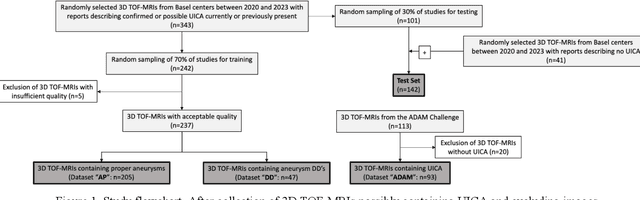
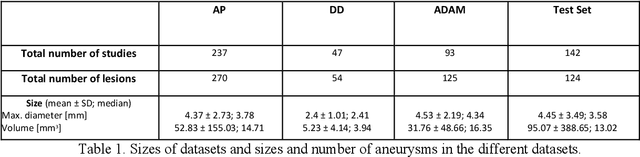
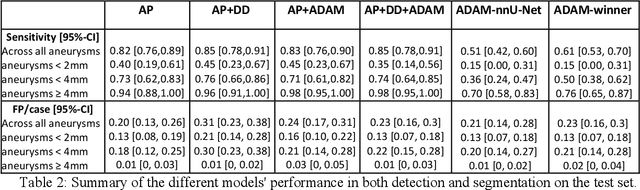
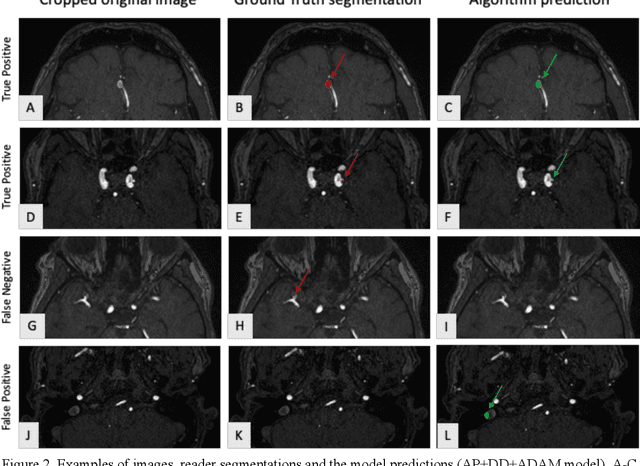
Abstract:Purpose: To develop an open-source nnU-Net-based AI model for combined detection and segmentation of unruptured intracranial aneurysms (UICA) in 3D TOF-MRI, and compare models trained on datasets with aneurysm-like differential diagnoses. Methods: This retrospective study (2020-2023) included 385 anonymized 3D TOF-MRI images from 364 patients (mean age 59 years, 60% female) at multiple centers plus 113 subjects from the ADAM challenge. Images featured untreated or possible UICAs and differential diagnoses. Four distinct training datasets were created, and the nnU-Net framework was used for model development. Performance was assessed on a separate test set using sensitivity and False Positive (FP)/case rate for detection, and DICE score and NSD (Normalized Surface Distance) with a 0.5mm threshold for segmentation. Statistical analysis included chi-square, Mann-Whitney-U, and Kruskal-Wallis tests, with significance set at p < 0.05. Results: Models achieved overall sensitivity between 82% and 85% and a FP/case rate of 0.20 to 0.31, with no significant differences (p = 0.90 and p = 0.16). The primary model showed 85% sensitivity and 0.23 FP/case rate, outperforming the ADAM-challenge winner (61%) and a nnU-Net trained on ADAM data (51%) in sensitivity (p < 0.05). It achieved a mean DICE score of 0.73 and an NSD of 0.84 for correctly detected UICA. Conclusions: Our open-source, nnU-Net-based AI model (available at 10.5281/zenodo.13386859) demonstrates high sensitivity, low false positive rates, and consistent segmentation accuracy for UICA detection and segmentation in 3D TOF-MRI, suggesting its potential to improve clinical diagnosis and for monitoring of UICA.
TotalSegmentator MRI: Sequence-Independent Segmentation of 59 Anatomical Structures in MR images
May 29, 2024



Abstract:Purpose: To develop an open-source and easy-to-use segmentation model that can automatically and robustly segment most major anatomical structures in MR images independently of the MR sequence. Materials and Methods: In this study we extended the capabilities of TotalSegmentator to MR images. 298 MR scans and 227 CT scans were used to segment 59 anatomical structures (20 organs, 18 bones, 11 muscles, 7 vessels, 3 tissue types) relevant for use cases such as organ volumetry, disease characterization, and surgical planning. The MR and CT images were randomly sampled from routine clinical studies and thus represent a real-world dataset (different ages, pathologies, scanners, body parts, sequences, contrasts, echo times, repetition times, field strengths, slice thicknesses and sites). We trained an nnU-Net segmentation algorithm on this dataset and calculated Dice similarity coefficients (Dice) to evaluate the model's performance. Results: The model showed a Dice score of 0.824 (CI: 0.801, 0.842) on the test set, which included a wide range of clinical data with major pathologies. The model significantly outperformed two other publicly available segmentation models (Dice score, 0.824 versus 0.762; p<0.001 and 0.762 versus 0.542; p<0.001). On the CT image test set of the original TotalSegmentator paper it almost matches the performance of the original TotalSegmentator (Dice score, 0.960 versus 0.970; p<0.001). Conclusion: Our proposed model extends the capabilities of TotalSegmentator to MR images. The annotated dataset (https://zenodo.org/doi/10.5281/zenodo.11367004) and open-source toolkit (https://www.github.com/wasserth/TotalSegmentator) are publicly available.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
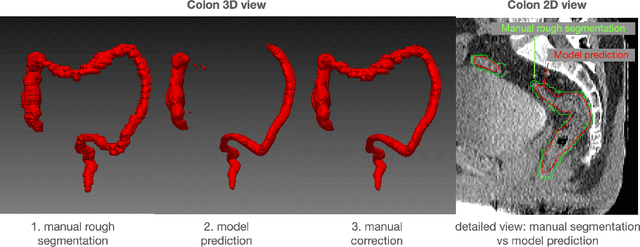
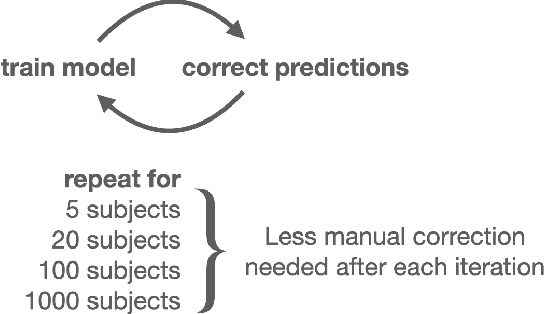
Deep Anatomical Federated Network (Dafne): an open client/server framework for the continuous collaborative improvement of deep-learning-based medical image segmentation
Feb 14, 2023Abstract:Semantic segmentation is a crucial step to extract quantitative information from medical (and, specifically, radiological) images to aid the diagnostic process, clinical follow-up. and to generate biomarkers for clinical research. In recent years, machine learning algorithms have become the primary tool for this task. However, its real-world performance is heavily reliant on the comprehensiveness of training data. Dafne is the first decentralized, collaborative solution that implements continuously evolving deep learning models exploiting the collective knowledge of the users of the system. In the Dafne workflow, the result of each automated segmentation is refined by the user through an integrated interface, so that the new information is used to continuously expand the training pool via federated incremental learning. The models deployed through Dafne are able to improve their performance over time and to generalize to data types not seen in the training sets, thus becoming a viable and practical solution for real-life medical segmentation tasks.
TotalSegmentator: robust segmentation of 104 anatomical structures in CT images
Aug 11, 2022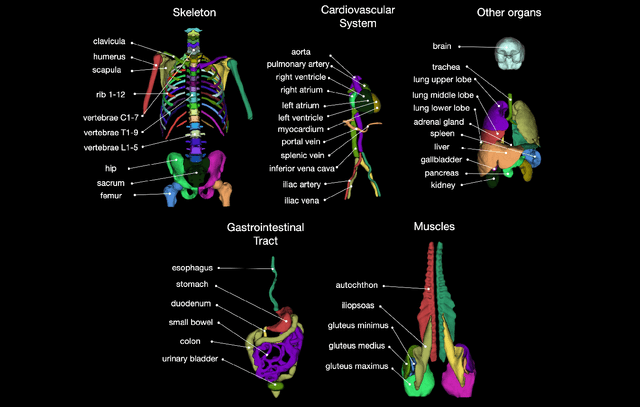
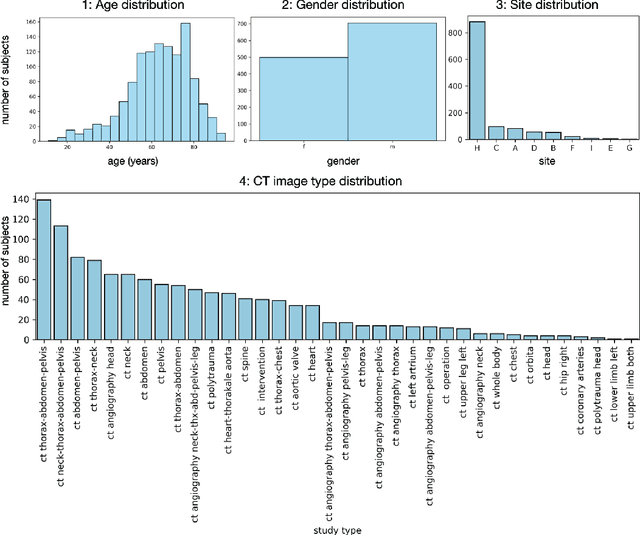
Abstract:In this work we focus on automatic segmentation of multiple anatomical structures in (whole body) CT images. Many segmentation algorithms exist for this task. However, in most cases they suffer from 3 problems: 1. They are difficult to use (the code and data is not publicly available or difficult to use). 2. They do not generalize (often the training dataset was curated to only contain very clean images which do not reflect the image distribution found during clinical routine), 3. The algorithm can only segment one anatomical structure. For more structures several algorithms have to be used which increases the effort required to set up the system. In this work we publish a new dataset and segmentation toolkit which solves all three of these problems: In 1204 CT images we segmented 104 anatomical structures (27 organs, 59 bones, 10 muscles, 8 vessels) covering a majority of relevant classes for most use cases. We show an improved workflow for the creation of ground truth segmentations which speeds up the process by over 10x. The CT images were randomly sampled from clinical routine, thus representing a real world dataset which generalizes to clinical application. The dataset contains a wide range of different pathologies, scanners, sequences and sites. Finally, we train a segmentation algorithm on this new dataset. We call this algorithm TotalSegmentator and make it easily available as a pretrained python pip package (pip install totalsegmentator). Usage is as simple as TotalSegmentator -i ct.nii.gz -o seg and it works well for most CT images. The code is available at https://github.com/wasserth/TotalSegmentator and the dataset at https://doi.org/10.5281/zenodo.6802613.
Combined tract segmentation and orientation mapping for bundle-specific tractography
Jan 29, 2019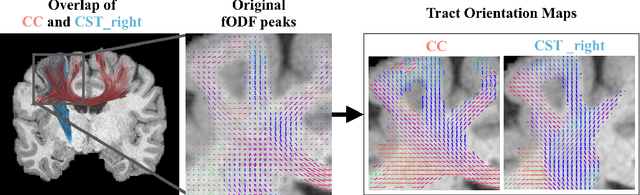
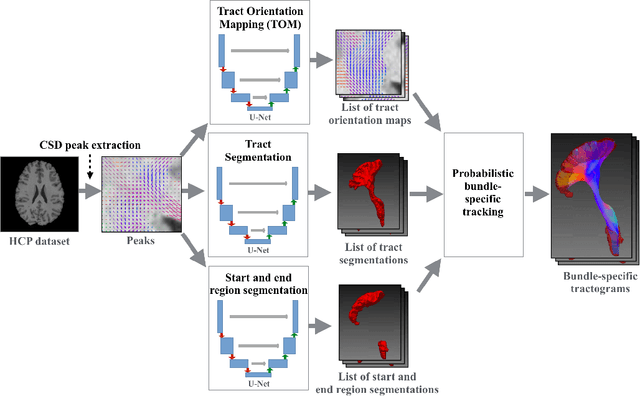
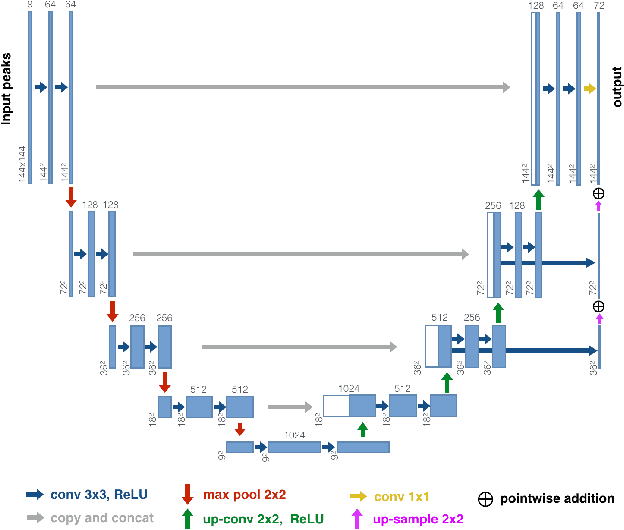
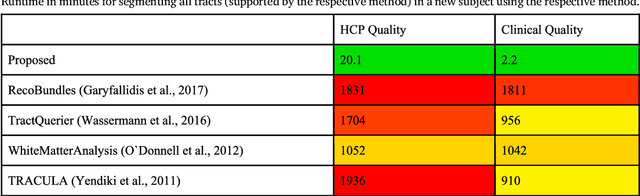
Abstract:While the major white matter tracts are of great interest to numerous studies in neuroscience and medicine, their manual dissection in larger cohorts from diffusion MRI tractograms is time-consuming, requires expert knowledge and is hard to reproduce. In previous work we presented tract orientation mapping (TOM) as a novel concept for bundle-specific tractography. It is based on a learned mapping from the original fiber orientation distribution function (fODF) peaks to tract specific peaks, called tract orientation maps. Each tract orientation map represents the voxel-wise principal orientation of one tract.Here, we present an extension of this approach that combines TOM with accurate segmentations of the tract outline and its start and end region. We also introduce a custom probabilistic tracking algorithm that samples from a Gaussian distribution with fixed standard deviation centered on each peak thus enabling more complete trackings on the tract orientation maps than deterministic tracking. These extensions enable the automatic creation of bundle-specific tractograms with previously unseen accuracy. We show for 72 different bundles on high quality, low quality and phantom data that our approach runs faster and produces more accurate bundle-specific tractograms than 7 state of the art benchmark methods while avoiding cumbersome processing steps like whole brain tractography, non-linear registration, clustering or manual dissection. Moreover, we show on 17 datasets that our approach generalizes well to datasets acquired with different scanners and settings as well as with pathologies. The code of our method is openly available at www.github.com/MIC-DKFZ/TractSeg.
nnU-Net: Self-adapting Framework for U-Net-Based Medical Image Segmentation
Sep 27, 2018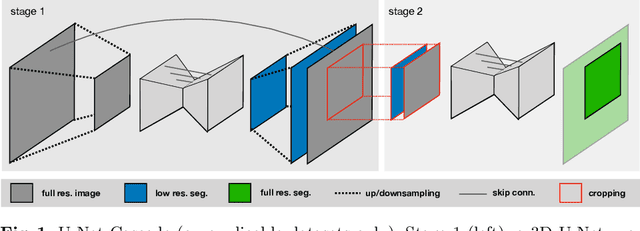
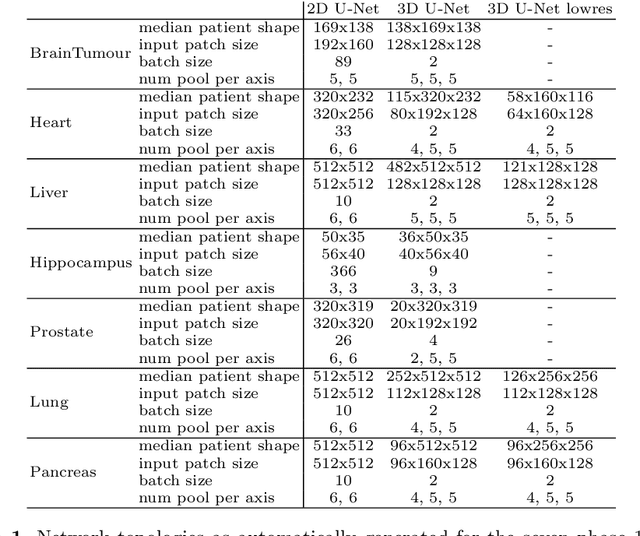
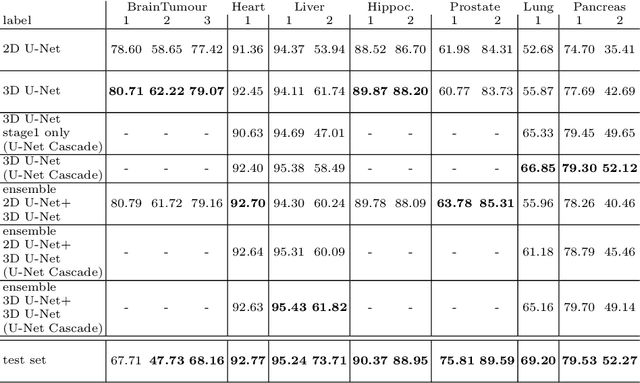
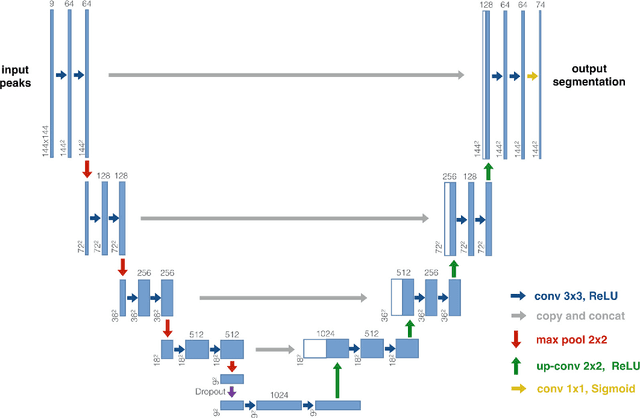
Abstract:The U-Net was presented in 2015. With its straight-forward and successful architecture it quickly evolved to a commonly used benchmark in medical image segmentation. The adaptation of the U-Net to novel problems, however, comprises several degrees of freedom regarding the exact architecture, preprocessing, training and inference. These choices are not independent of each other and substantially impact the overall performance. The present paper introduces the nnU-Net ('no-new-Net'), which refers to a robust and self-adapting framework on the basis of 2D and 3D vanilla U-Nets. We argue the strong case for taking away superfluous bells and whistles of many proposed network designs and instead focus on the remaining aspects that make out the performance and generalizability of a method. We evaluate the nnU-Net in the context of the Medical Segmentation Decathlon challenge, which measures segmentation performance in ten disciplines comprising distinct entities, image modalities, image geometries and dataset sizes, with no manual adjustments between datasets allowed. At the time of manuscript submission, nnU-Net achieves the highest mean dice scores across all classes and seven phase 1 tasks (except class 1 in BrainTumour) in the online leaderboard of the challenge.
TractSeg - Fast and accurate white matter tract segmentation
Aug 20, 2018
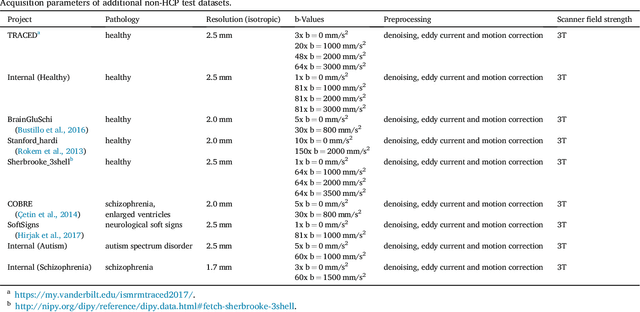
Abstract:The individual course of white matter fiber tracts is an important key for analysis of white matter characteristics in healthy and diseased brains. Uniquely, diffusion-weighted MRI tractography in combination with region-based or clustering-based selection of streamlines allows for the in-vivo delineation and analysis of anatomically well known tracts. This, however, currently requires complex, computationally intensive and tedious-to-set-up processing pipelines. TractSeg is a novel convolutional neural network-based approach that directly segments tracts in the field of fiber orientation distribution function (fODF) peaks without requiring tractography, image registration or parcellation. We demonstrate in 105 subjects from the Human Connectome Project that the proposed approach is much faster than existing methods while providing unprecedented accuracy. The code and data are openly available at https://github.com/MIC-DKFZ/TractSeg/ and https://doi.org/10.5281/zenodo.1088277, respectively.
Tract orientation mapping for bundle-specific tractography
Jun 14, 2018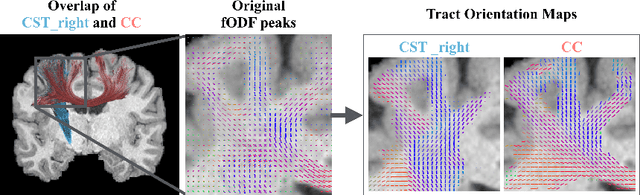
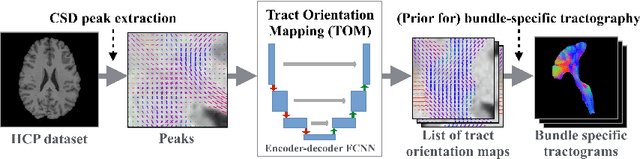
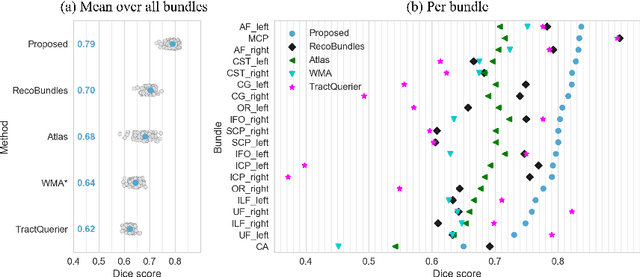
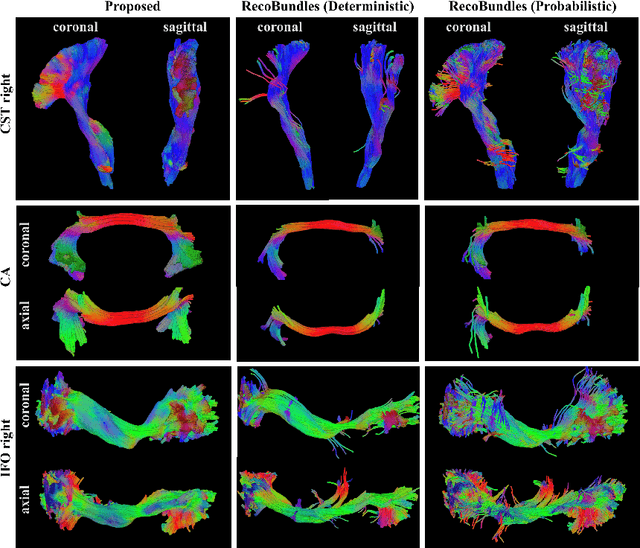
Abstract:While the major white matter tracts are of great interest to numerous studies in neuroscience and medicine, their manual dissection in larger cohorts from diffusion MRI tractograms is time-consuming, requires expert knowledge and is hard to reproduce. Tract orientation mapping (TOM) is a novel concept that facilitates bundle-specific tractography based on a learned mapping from the original fiber orientation distribution function (fODF) peaks to a list of tract orientation maps (also abbr. TOM). Each TOM represents one of the known tracts with each voxel containing no more than one orientation vector. TOMs can act as a prior or even as direct input for tractography. We use an encoder-decoder fully-convolutional neural network architecture to learn the required mapping. In comparison to previous concepts for the reconstruction of specific bundles, the presented one avoids various cumbersome processing steps like whole brain tractography, atlas registration or clustering. We compare it to four state of the art bundle recognition methods on 20 different bundles in a total of 105 subjects from the Human Connectome Project. Results are anatomically convincing even for difficult tracts, while reaching low angular errors, unprecedented runtimes and top accuracy values (Dice). Our code and our data are openly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge