Marco Reisert
TotalVibeSegmentator: Full Torso Segmentation for the NAKO and UK Biobank in Volumetric Interpolated Breath-hold Examination Body Images
May 31, 2024



Abstract:Objectives: To present a publicly available torso segmentation network for large epidemiology datasets on volumetric interpolated breath-hold examination (VIBE) images. Materials & Methods: We extracted preliminary segmentations from TotalSegmentator, spine, and body composition networks for VIBE images, then improved them iteratively and retrained a nnUNet network. Using subsets of NAKO (85 subjects) and UK Biobank (16 subjects), we evaluated with Dice-score on a holdout set (12 subjects) and existing organ segmentation approach (1000 subjects), generating 71 semantic segmentation types for VIBE images. We provide an additional network for the vertebra segments 22 individual vertebra types. Results: We achieved an average Dice score of 0.89 +- 0.07 overall 71 segmentation labels. We scored > 0.90 Dice-score on the abdominal organs except for the pancreas with a Dice of 0.70. Conclusion: Our work offers a detailed and refined publicly available full torso segmentation on VIBE images.
TotalSegmentator MRI: Sequence-Independent Segmentation of 59 Anatomical Structures in MR images
May 29, 2024



Abstract:Purpose: To develop an open-source and easy-to-use segmentation model that can automatically and robustly segment most major anatomical structures in MR images independently of the MR sequence. Materials and Methods: In this study we extended the capabilities of TotalSegmentator to MR images. 298 MR scans and 227 CT scans were used to segment 59 anatomical structures (20 organs, 18 bones, 11 muscles, 7 vessels, 3 tissue types) relevant for use cases such as organ volumetry, disease characterization, and surgical planning. The MR and CT images were randomly sampled from routine clinical studies and thus represent a real-world dataset (different ages, pathologies, scanners, body parts, sequences, contrasts, echo times, repetition times, field strengths, slice thicknesses and sites). We trained an nnU-Net segmentation algorithm on this dataset and calculated Dice similarity coefficients (Dice) to evaluate the model's performance. Results: The model showed a Dice score of 0.824 (CI: 0.801, 0.842) on the test set, which included a wide range of clinical data with major pathologies. The model significantly outperformed two other publicly available segmentation models (Dice score, 0.824 versus 0.762; p<0.001 and 0.762 versus 0.542; p<0.001). On the CT image test set of the original TotalSegmentator paper it almost matches the performance of the original TotalSegmentator (Dice score, 0.960 versus 0.970; p<0.001). Conclusion: Our proposed model extends the capabilities of TotalSegmentator to MR images. The annotated dataset (https://zenodo.org/doi/10.5281/zenodo.11367004) and open-source toolkit (https://www.github.com/wasserth/TotalSegmentator) are publicly available.
PatchMorph: A Stochastic Deep Learning Approach for Unsupervised 3D Brain Image Registration with Small Patches
Dec 12, 2023Abstract:We introduce "PatchMorph," an new stochastic deep learning algorithm tailored for unsupervised 3D brain image registration. Unlike other methods, our method uses compact patches of a constant small size to derive solutions that can combine global transformations with local deformations. This approach minimizes the memory footprint of the GPU during training, but also enables us to operate on numerous amounts of randomly overlapping small patches during inference to mitigate image and patch boundary problems. PatchMorph adeptly handles world coordinate transformations between two input images, accommodating variances in attributes such as spacing, array sizes, and orientations. The spatial resolution of patches transitions from coarse to fine, addressing both global and local attributes essential for aligning the images. Each patch offers a unique perspective, together converging towards a comprehensive solution. Experiments on human T1 MRI brain images and marmoset brain images from serial 2-photon tomography affirm PatchMorph's superior performance.
Deep Neural Patchworks: Coping with Large Segmentation Tasks
Jun 07, 2022


Abstract:Convolutional neural networks are the way to solve arbitrary image segmentation tasks. However, when images are large, memory demands often exceed the available resources, in particular on a common GPU. Especially in biomedical imaging, where 3D images are common, the problems are apparent. A typical approach to solve this limitation is to break the task into smaller subtasks by dividing images into smaller image patches. Another approach, if applicable, is to look at the 2D image sections separately, and to solve the problem in 2D. Often, the loss of global context makes such approaches less effective; important global information might not be present in the current image patch, or the selected 2D image section. Here, we propose Deep Neural Patchworks (DNP), a segmentation framework that is based on hierarchical and nested stacking of patch-based networks that solves the dilemma between global context and memory limitations.
HAMLET: Hierarchical Harmonic Filters for Learning Tracts from Diffusion MRI
Jul 03, 2018


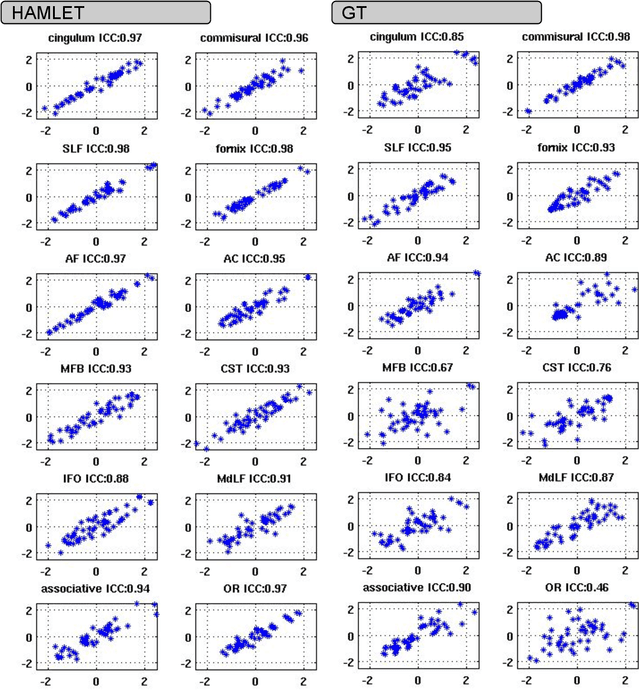
Abstract:In this work we propose HAMLET, a novel tract learning algorithm, which, after training, maps raw diffusion weighted MRI directly onto an image which simultaneously indicates tract direction and tract presence. The automatic learning of fiber tracts based on diffusion MRI data is a rather new idea, which tries to overcome limitations of atlas-based techniques. HAMLET takes a such an approach. Unlike the current trend in machine learning, HAMLET has only a small number of free parameters HAMLET is based on spherical tensor algebra which allows a translation and rotation covariant treatment of the problem. HAMLET is based on a repeated application of convolutions and non-linearities, which all respect the rotation covariance. The intrinsic treatment of such basic image transformations in HAMLET allows the training and generalization of the algorithm without any additional data augmentation. We demonstrate the performance of our approach for twelve prominent bundles, and show that the obtained tract estimates are robust and reliable. It is also shown that the learned models are portable from one sequence to another.
Gibbs-Ringing Artifact Removal Based on Local Subvoxel-shifts
Jan 30, 2015



Abstract:Gibbs-ringing is a well known artifact which manifests itself as spurious oscillations in the vicinity of sharp image transients, e.g. at tissue boundaries. The origin can be seen in the truncation of k-space during MRI data-acquisition. Consequently, correction techniques like Gegenbauer reconstruction or extrapolation methods aim at recovering these missing data. Here, we present a simple and robust method which exploits a different view on the Gibbs-phenomena. The truncation in k-space can be interpreted as a convolution with a sinc-function in image space. Hence, the severity of the artifacts depends on how the sinc-function is sampled. We propose to re-interpolate the image based on local, subvoxel shifts to sample the ringing pattern at the zero-crossings of the oscillating sinc-function. With this, the artifact can effectively and robustly be removed with a minimal amount of smoothing.
Left-Invariant Diffusion on the Motion Group in terms of the Irreducible Representations of SO
Feb 24, 2012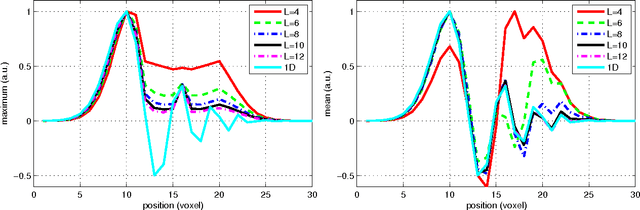
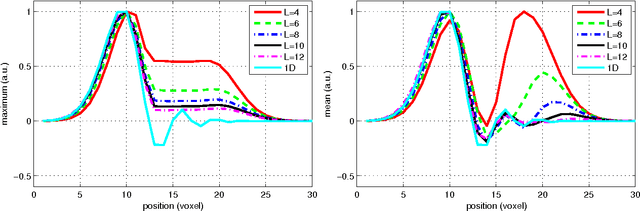


Abstract:In this work we study the formulation of convection/diffusion equations on the 3D motion group SE(3) in terms of the irreducible representations of SO(3). Therefore, the left-invariant vector-fields on SE(3) are expressed as linear operators, that are differential forms in the translation coordinate and algebraic in the rotation. In the context of 3D image processing this approach avoids the explicit discretization of SO(3) or $S_2$, respectively. This is particular important for SO(3), where a direct discretization is infeasible due to the enormous memory consumption. We show two applications of the framework: one in the context of diffusion-weighted magnetic resonance imaging and one in the context of object detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge