Michal Byra
Routing the Lottery: Adaptive Subnetworks for Heterogeneous Data
Jan 29, 2026Abstract:In pruning, the Lottery Ticket Hypothesis posits that large networks contain sparse subnetworks, or winning tickets, that can be trained in isolation to match the performance of their dense counterparts. However, most existing approaches assume a single universal winning ticket shared across all inputs, ignoring the inherent heterogeneity of real-world data. In this work, we propose Routing the Lottery (RTL), an adaptive pruning framework that discovers multiple specialized subnetworks, called adaptive tickets, each tailored to a class, semantic cluster, or environmental condition. Across diverse datasets and tasks, RTL consistently outperforms single- and multi-model baselines in balanced accuracy and recall, while using up to 10 times fewer parameters than independent models and exhibiting semantically aligned. Furthermore, we identify subnetwork collapse, a performance drop under aggressive pruning, and introduce a subnetwork similarity score that enables label-free diagnosis of oversparsification. Overall, our results recast pruning as a mechanism for aligning model structure with data heterogeneity, paving the way toward more modular and context-aware deep learning.
Generating visual explanations from deep networks using implicit neural representations
Jan 20, 2025



Abstract:Explaining deep learning models in a way that humans can easily understand is essential for responsible artificial intelligence applications. Attribution methods constitute an important area of explainable deep learning. The attribution problem involves finding parts of the network's input that are the most responsible for the model's output. In this work, we demonstrate that implicit neural representations (INRs) constitute a good framework for generating visual explanations. Firstly, we utilize coordinate-based implicit networks to reformulate and extend the extremal perturbations technique and generate attribution masks. Experimental results confirm the usefulness of our method. For instance, by proper conditioning of the implicit network, we obtain attribution masks that are well-behaved with respect to the imposed area constraints. Secondly, we present an iterative INR-based method that can be used to generate multiple non-overlapping attribution masks for the same image. We depict that a deep learning model may associate the image label with both the appearance of the object of interest as well as with areas and textures usually accompanying the object. Our study demonstrates that implicit networks are well-suited for the generation of attribution masks and can provide interesting insights about the performance of deep learning models.
Implicit Neural Representations for Speed-of-Sound Estimation in Ultrasound
Sep 21, 2024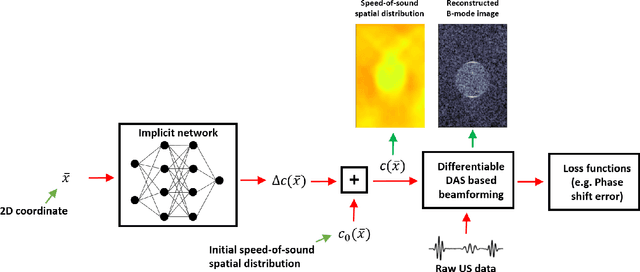
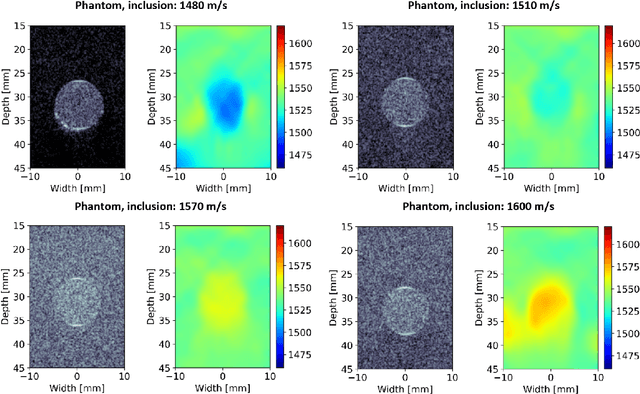
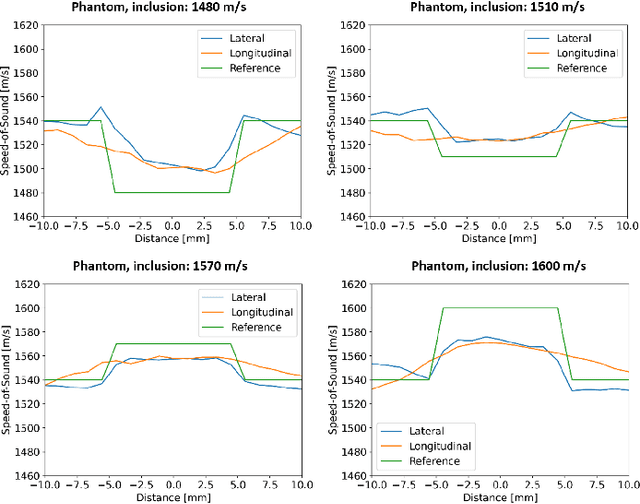
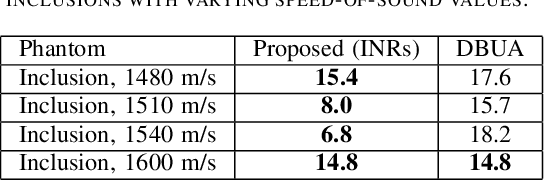
Abstract:Accurate estimation of the speed-of-sound (SoS) is important for ultrasound (US) image reconstruction techniques and tissue characterization. Various approaches have been proposed to calculate SoS, ranging from tomography-inspired algorithms like CUTE to convolutional networks, and more recently, physics-informed optimization frameworks based on differentiable beamforming. In this work, we utilize implicit neural representations (INRs) for SoS estimation in US. INRs are a type of neural network architecture that encodes continuous functions, such as images or physical quantities, through the weights of a network. Implicit networks may overcome the current limitations of SoS estimation techniques, which mainly arise from the use of non-adaptable and oversimplified physical models of tissue. Moreover, convolutional networks for SoS estimation, usually trained using simulated data, often fail when applied to real tissues due to out-of-distribution and data-shift issues. In contrast, implicit networks do not require extensive training datasets since each implicit network is optimized for an individual data case. This adaptability makes them suitable for processing US data collected from varied tissues and across different imaging protocols. We evaluated the proposed SoS estimation method based on INRs using data collected from a tissue-mimicking phantom containing four cylindrical inclusions, with SoS values ranging from 1480 m/s to 1600 m/s. The inclusions were immersed in a material with an SoS value of 1540 m/s. In experiments, the proposed method achieved strong performance, clearly demonstrating the usefulness of implicit networks for quantitative US applications.
PatchMorph: A Stochastic Deep Learning Approach for Unsupervised 3D Brain Image Registration with Small Patches
Dec 12, 2023Abstract:We introduce "PatchMorph," an new stochastic deep learning algorithm tailored for unsupervised 3D brain image registration. Unlike other methods, our method uses compact patches of a constant small size to derive solutions that can combine global transformations with local deformations. This approach minimizes the memory footprint of the GPU during training, but also enables us to operate on numerous amounts of randomly overlapping small patches during inference to mitigate image and patch boundary problems. PatchMorph adeptly handles world coordinate transformations between two input images, accommodating variances in attributes such as spacing, array sizes, and orientations. The spatial resolution of patches transitions from coarse to fine, addressing both global and local attributes essential for aligning the images. Each patch offers a unique perspective, together converging towards a comprehensive solution. Experiments on human T1 MRI brain images and marmoset brain images from serial 2-photon tomography affirm PatchMorph's superior performance.
Few-shot medical image classification with simple shape and texture text descriptors using vision-language models
Aug 08, 2023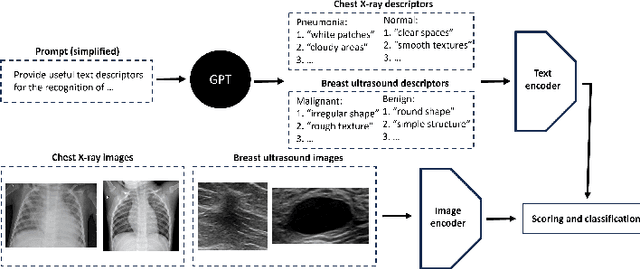
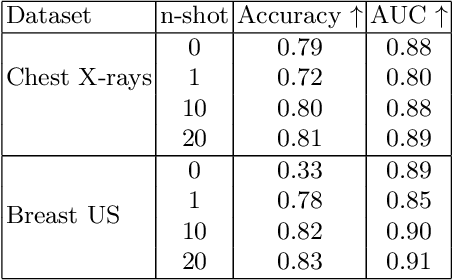
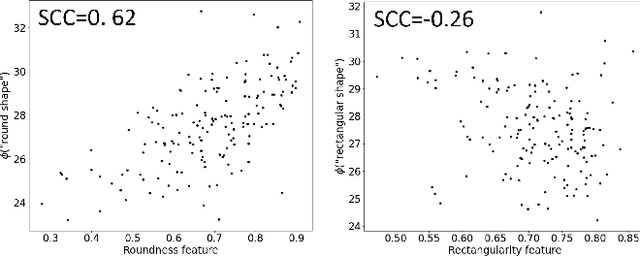

Abstract:In this work, we investigate the usefulness of vision-language models (VLMs) and large language models for binary few-shot classification of medical images. We utilize the GPT-4 model to generate text descriptors that encapsulate the shape and texture characteristics of objects in medical images. Subsequently, these GPT-4 generated descriptors, alongside VLMs pre-trained on natural images, are employed to classify chest X-rays and breast ultrasound images. Our results indicate that few-shot classification of medical images using VLMs and GPT-4 generated descriptors is a viable approach. However, accurate classification requires to exclude certain descriptors from the calculations of the classification scores. Moreover, we assess the ability of VLMs to evaluate shape features in breast mass ultrasound images. We further investigate the degree of variability among the sets of text descriptors produced by GPT-4. Our work provides several important insights about the application of VLMs for medical image analysis.
Implicit neural representations for joint decomposition and registration of gene expression images in the marmoset brain
Aug 08, 2023Abstract:We propose a novel image registration method based on implicit neural representations that addresses the challenging problem of registering a pair of brain images with similar anatomical structures, but where one image contains additional features or artifacts that are not present in the other image. To demonstrate its effectiveness, we use 2D microscopy $\textit{in situ}$ hybridization gene expression images of the marmoset brain. Accurately quantifying gene expression requires image registration to a brain template, which is difficult due to the diversity of patterns causing variations in visible anatomical brain structures. Our approach uses implicit networks in combination with an image exclusion loss to jointly perform the registration and decompose the image into a support and residual image. The support image aligns well with the template, while the residual image captures individual image characteristics that diverge from the template. In experiments, our method provided excellent results and outperformed other registration techniques.
An automated pipeline to create an atlas of in situ hybridization gene expression data in the adult marmoset brain
Mar 13, 2023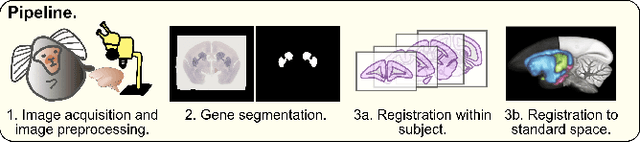
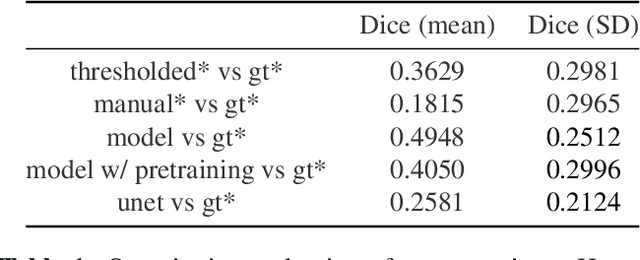
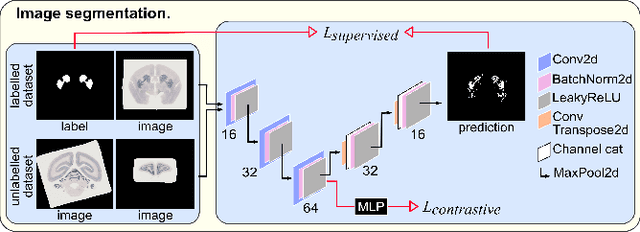

Abstract:We present the first automated pipeline to create an atlas of in situ hybridization gene expression in the adult marmoset brain in the same stereotaxic space. The pipeline consists of segmentation of gene expression from microscopy images and registration of images to a standard space. Automation of this pipeline is necessary to analyze the large volume of data in the genome-wide whole-brain dataset, and to process images that have varying intensity profiles and expression patterns with minimal human bias. To reduce the number of labelled images required for training, we develop a semi-supervised segmentation model. We further develop an iterative algorithm to register images to a standard space, enabling comparative analysis between genes and concurrent visualization with other datasets, thereby facilitating a more holistic understanding of primate brain structure and function.
Deep meta-learning for the selection of accurate ultrasound based breast mass classifier
Nov 03, 2022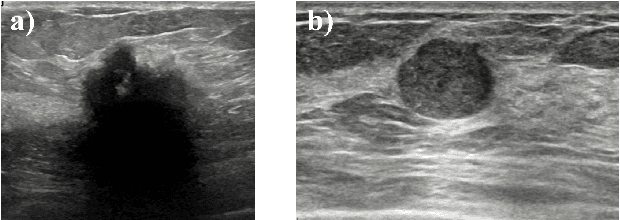
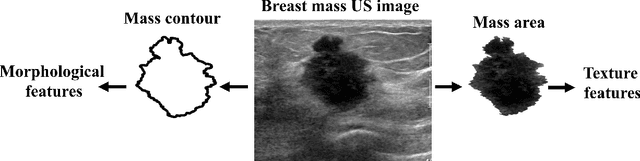
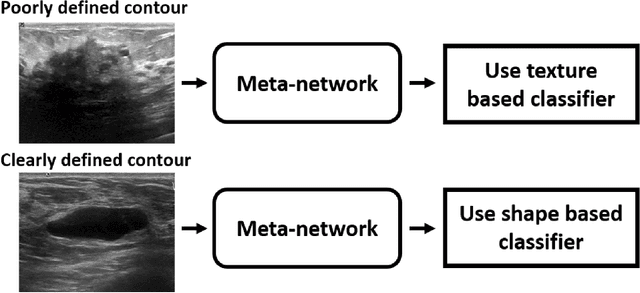
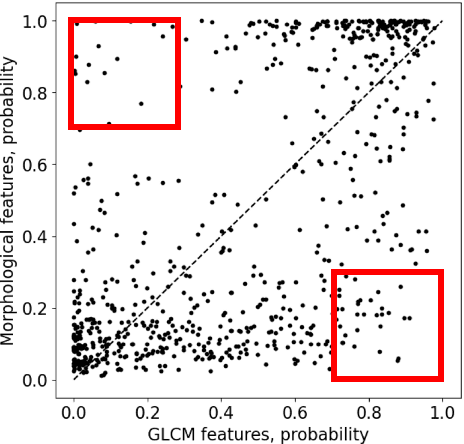
Abstract:Standard classification methods based on handcrafted morphological and texture features have achieved good performance in breast mass differentiation in ultrasound (US). In comparison to deep neural networks, commonly perceived as "black-box" models, classical techniques are based on features that have well-understood medical and physical interpretation. However, classifiers based on morphological features commonly underperform in the presence of the shadowing artifact and ill-defined mass borders, while texture based classifiers may fail when the US image is too noisy. Therefore, in practice it would be beneficial to select the classification method based on the appearance of the particular US image. In this work, we develop a deep meta-network that can automatically process input breast mass US images and recommend whether to apply the shape or texture based classifier for the breast mass differentiation. Our preliminary results demonstrate that meta-learning techniques can be used to improve the performance of the standard classifiers based on handcrafted features. With the proposed meta-learning based approach, we achieved the area under the receiver operating characteristic curve of 0.95 and accuracy of 0.91.
Parameter estimation of the homodyned K distribution based on neural networks and trainable fractional-order moments
Oct 11, 2022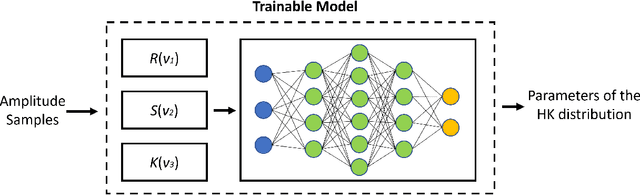
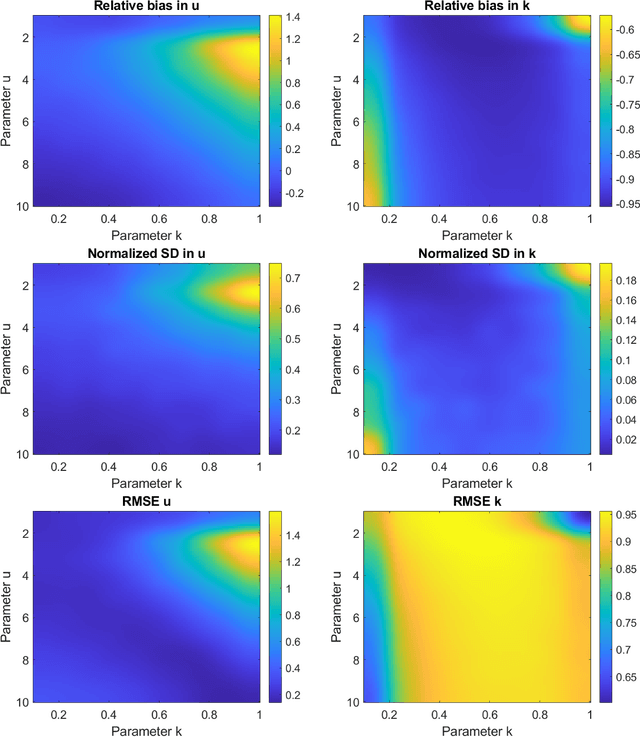
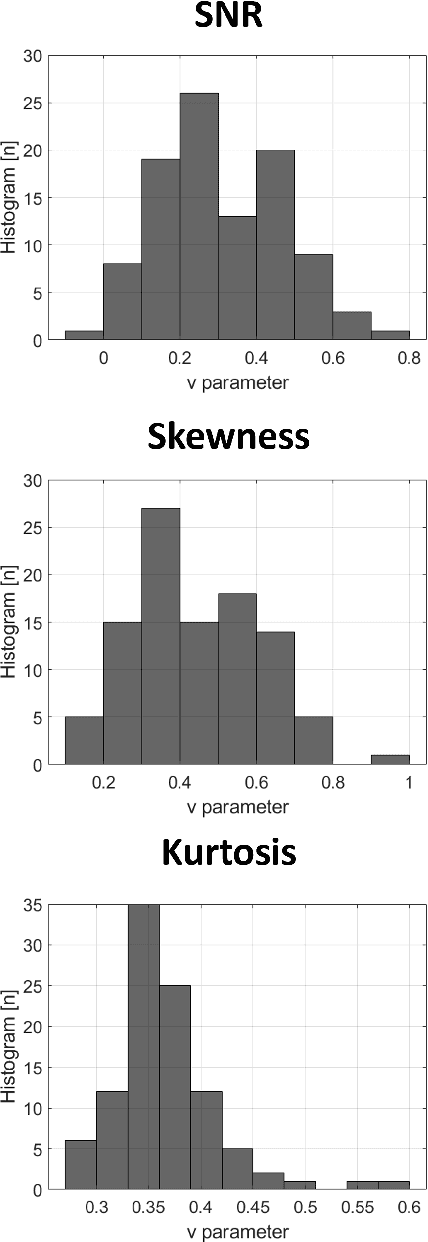
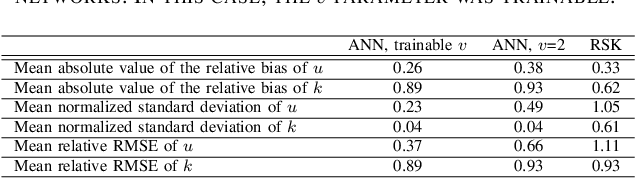
Abstract:Homodyned K (HK) distribution has been widely used to describe the scattering phenomena arising in various research fields, such as ultrasound imaging or optics. In this work, we propose a machine learning based approach to the estimation of the HK distribution parameters. We develop neural networks that can estimate the HK distribution parameters based on the signal-to-noise ratio, skewness and kurtosis calculated using fractional-order moments. Compared to the previous approaches, we consider the orders of the moments as trainable variables that can be optimized along with the network weights using the back-propagation algorithm. Networks are trained based on samples generated from the HK distribution. Obtained results demonstrate that the proposed method can be used to accurately estimate the HK distribution parameters.
Estimating the ultrasound attenuation coefficient using convolutional neural networks -- a feasibility study
May 19, 2022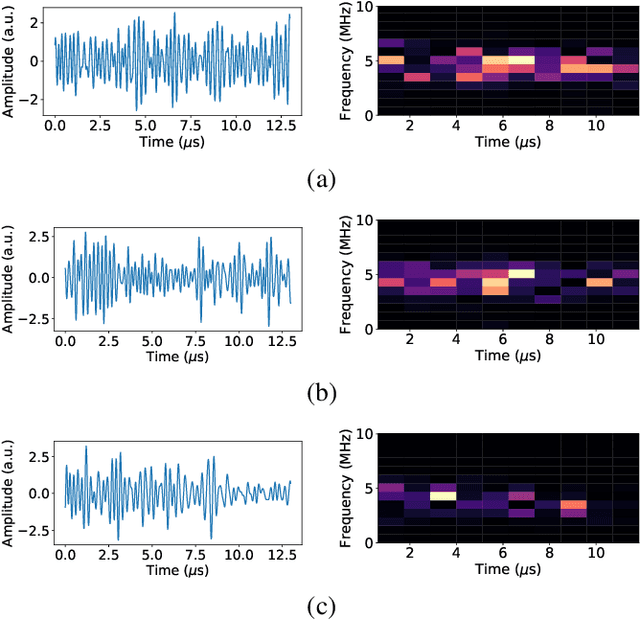
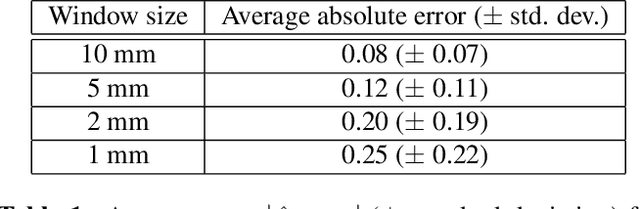
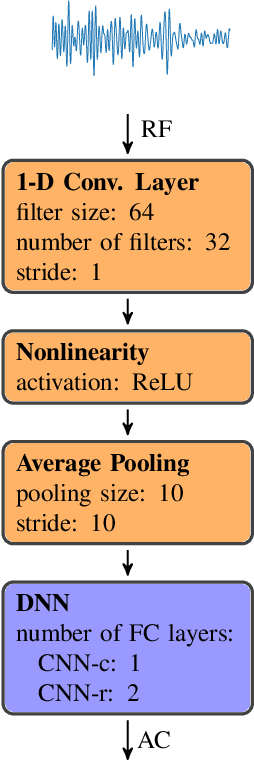
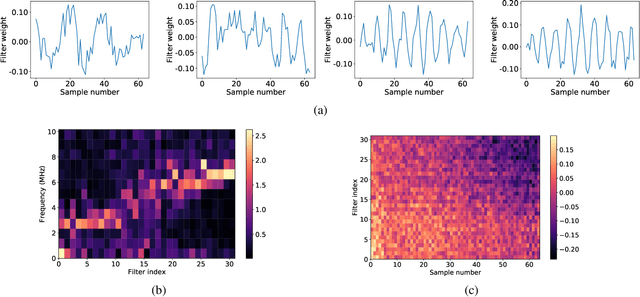
Abstract:Attenuation coefficient (AC) is a fundamental measure of tissue acoustical properties, which can be used in medical diagnostics. In this work, we investigate the feasibility of using convolutional neural networks (CNNs) to directly estimate AC from radio-frequency (RF) ultrasound signals. To develop the CNNs we used RF signals collected from tissue mimicking numerical phantoms for the AC values in a range from 0.1 to 1.5 dB/(MHz*cm). The models were trained based on 1-D patches of RF data. We obtained mean absolute AC estimation errors of 0.08, 0.12, 0.20, 0.25 for the patch lengths: 10 mm, 5 mm, 2 mm and 1 mm, respectively. We explain the performance of the model by visualizing the frequency content associated with convolutional filters. Our study presents that the AC can be calculated using deep learning, and the weights of the CNNs can have physical interpretation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge