Piotr Komorowski
Towards Visually Explaining Statistical Tests with Applications in Biomedical Imaging
Jan 20, 2026Abstract:Deep neural two-sample tests have recently shown strong power for detecting distributional differences between groups, yet their black-box nature limits interpretability and practical adoption in biomedical analysis. Moreover, most existing post-hoc explainability methods rely on class labels, making them unsuitable for label-free statistical testing settings. We propose an explainable deep statistical testing framework that augments deep two-sample tests with sample-level and feature-level explanations, revealing which individual samples and which input features drive statistically significant group differences. Our method highlights which image regions and which individual samples contribute most to the detected group difference, providing spatial and instance-wise insight into the test's decision. Applied to biomedical imaging data, the proposed framework identifies influential samples and highlights anatomically meaningful regions associated with disease-related variation. This work bridges statistical inference and explainable AI, enabling interpretable, label-free population analysis in medical imaging.
Aggregated Attributions for Explanatory Analysis of 3D Segmentation Models
Jul 24, 2024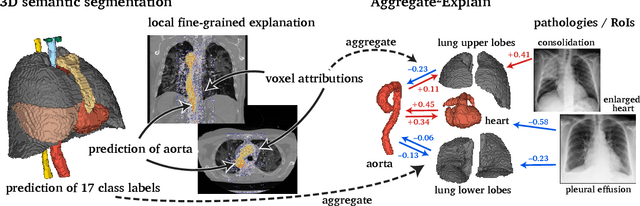
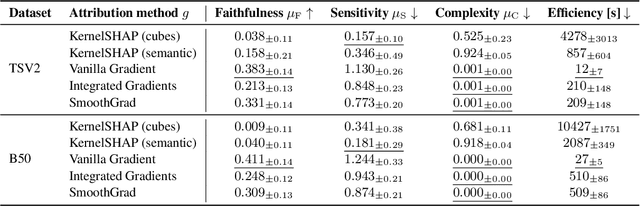
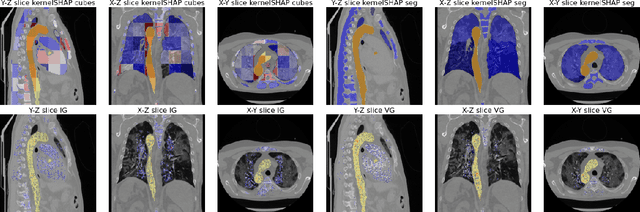
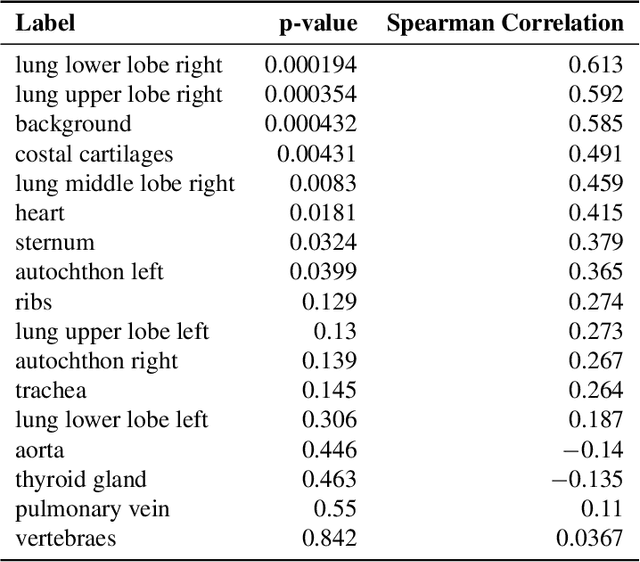
Abstract:Analysis of 3D segmentation models, especially in the context of medical imaging, is often limited to segmentation performance metrics that overlook the crucial aspect of explainability and bias. Currently, effectively explaining these models with saliency maps is challenging due to the high dimensions of input images multiplied by the ever-growing number of segmented class labels. To this end, we introduce Agg^2Exp, a methodology for aggregating fine-grained voxel attributions of the segmentation model's predictions. Unlike classical explanation methods that primarily focus on the local feature attribution, Agg^2Exp enables a more comprehensive global view on the importance of predicted segments in 3D images. Our benchmarking experiments show that gradient-based voxel attributions are more faithful to the model's predictions than perturbation-based explanations. As a concrete use-case, we apply Agg^2Exp to discover knowledge acquired by the Swin UNEt TRansformer model trained on the TotalSegmentator v2 dataset for segmenting anatomical structures in computed tomography medical images. Agg^2Exp facilitates the explanatory analysis of large segmentation models beyond their predictive performance.
Towards Evaluating Explanations of Vision Transformers for Medical Imaging
Apr 12, 2023Abstract:As deep learning models increasingly find applications in critical domains such as medical imaging, the need for transparent and trustworthy decision-making becomes paramount. Many explainability methods provide insights into how these models make predictions by attributing importance to input features. As Vision Transformer (ViT) becomes a promising alternative to convolutional neural networks for image classification, its interpretability remains an open research question. This paper investigates the performance of various interpretation methods on a ViT applied to classify chest X-ray images. We introduce the notion of evaluating faithfulness, sensitivity, and complexity of ViT explanations. The obtained results indicate that Layerwise relevance propagation for transformers outperforms Local interpretable model-agnostic explanations and Attention visualization, providing a more accurate and reliable representation of what a ViT has actually learned. Our findings provide insights into the applicability of ViT explanations in medical imaging and highlight the importance of using appropriate evaluation criteria for comparing them.
Deep meta-learning for the selection of accurate ultrasound based breast mass classifier
Nov 03, 2022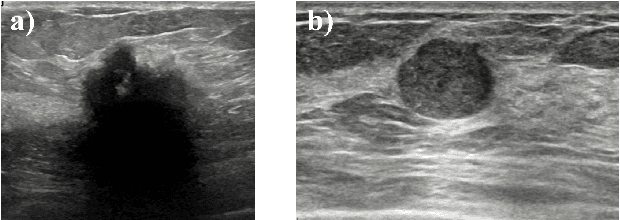
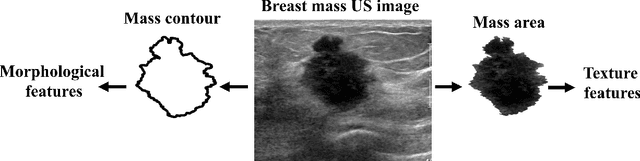
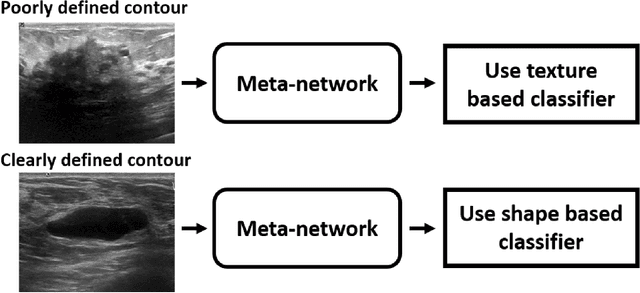
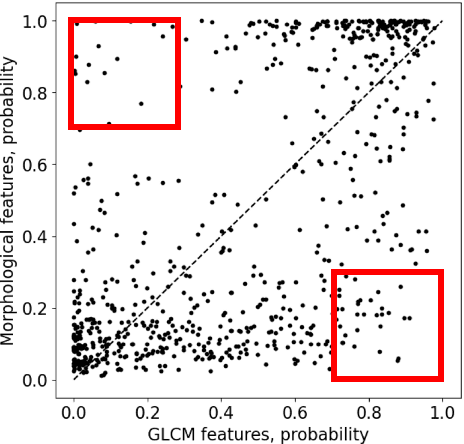
Abstract:Standard classification methods based on handcrafted morphological and texture features have achieved good performance in breast mass differentiation in ultrasound (US). In comparison to deep neural networks, commonly perceived as "black-box" models, classical techniques are based on features that have well-understood medical and physical interpretation. However, classifiers based on morphological features commonly underperform in the presence of the shadowing artifact and ill-defined mass borders, while texture based classifiers may fail when the US image is too noisy. Therefore, in practice it would be beneficial to select the classification method based on the appearance of the particular US image. In this work, we develop a deep meta-network that can automatically process input breast mass US images and recommend whether to apply the shape or texture based classifier for the breast mass differentiation. Our preliminary results demonstrate that meta-learning techniques can be used to improve the performance of the standard classifiers based on handcrafted features. With the proposed meta-learning based approach, we achieved the area under the receiver operating characteristic curve of 0.95 and accuracy of 0.91.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge