Ziemowit Klimonda
Implicit Neural Representations for Speed-of-Sound Estimation in Ultrasound
Sep 21, 2024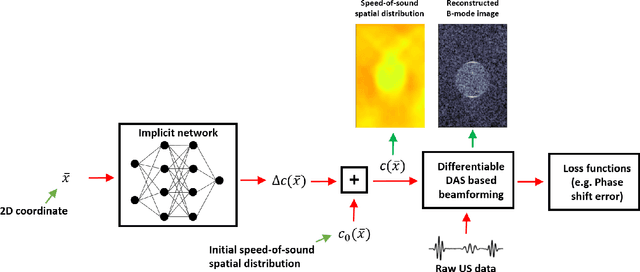
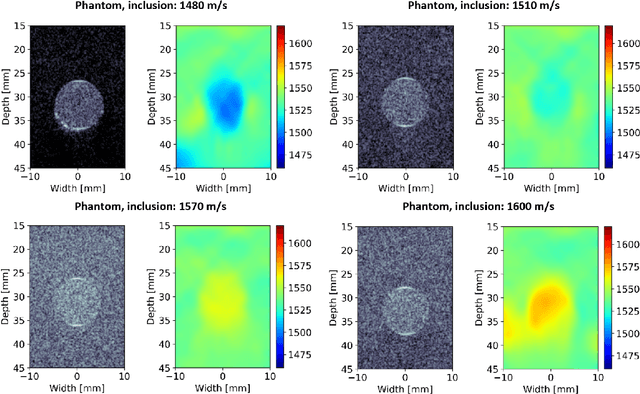
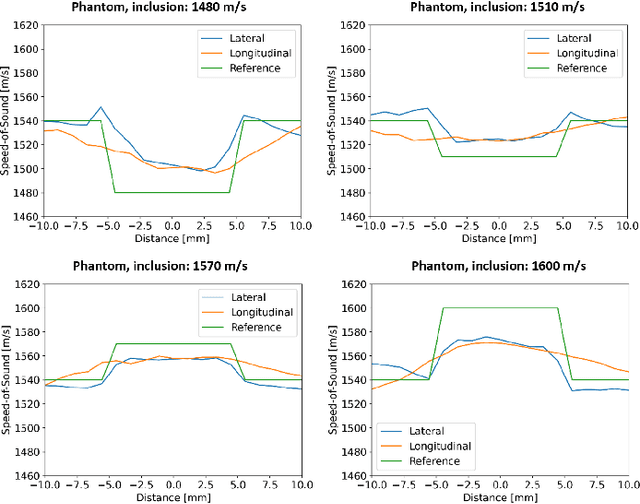
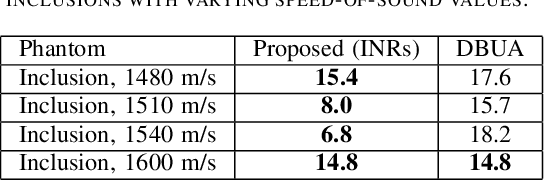
Abstract:Accurate estimation of the speed-of-sound (SoS) is important for ultrasound (US) image reconstruction techniques and tissue characterization. Various approaches have been proposed to calculate SoS, ranging from tomography-inspired algorithms like CUTE to convolutional networks, and more recently, physics-informed optimization frameworks based on differentiable beamforming. In this work, we utilize implicit neural representations (INRs) for SoS estimation in US. INRs are a type of neural network architecture that encodes continuous functions, such as images or physical quantities, through the weights of a network. Implicit networks may overcome the current limitations of SoS estimation techniques, which mainly arise from the use of non-adaptable and oversimplified physical models of tissue. Moreover, convolutional networks for SoS estimation, usually trained using simulated data, often fail when applied to real tissues due to out-of-distribution and data-shift issues. In contrast, implicit networks do not require extensive training datasets since each implicit network is optimized for an individual data case. This adaptability makes them suitable for processing US data collected from varied tissues and across different imaging protocols. We evaluated the proposed SoS estimation method based on INRs using data collected from a tissue-mimicking phantom containing four cylindrical inclusions, with SoS values ranging from 1480 m/s to 1600 m/s. The inclusions were immersed in a material with an SoS value of 1540 m/s. In experiments, the proposed method achieved strong performance, clearly demonstrating the usefulness of implicit networks for quantitative US applications.
Deep meta-learning for the selection of accurate ultrasound based breast mass classifier
Nov 03, 2022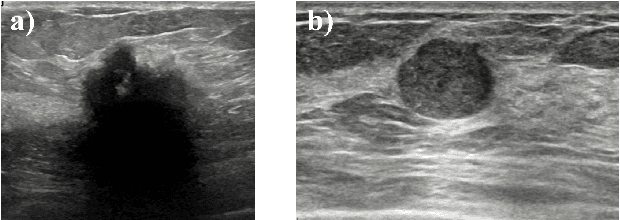
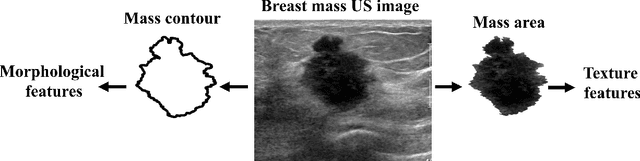
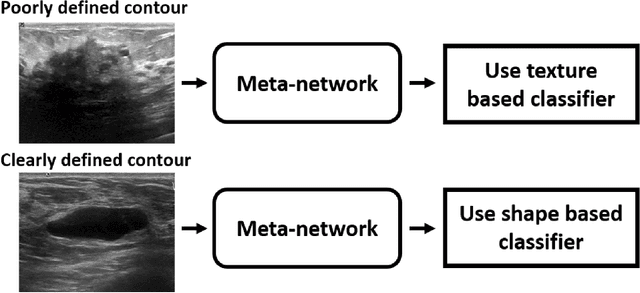
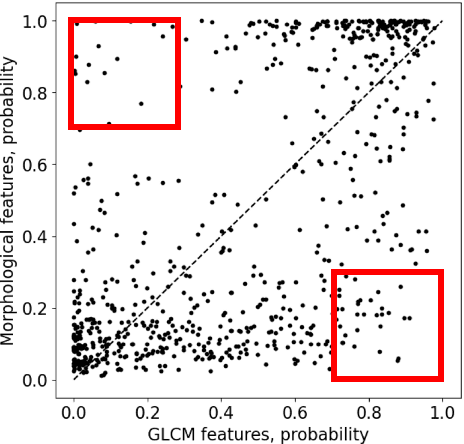
Abstract:Standard classification methods based on handcrafted morphological and texture features have achieved good performance in breast mass differentiation in ultrasound (US). In comparison to deep neural networks, commonly perceived as "black-box" models, classical techniques are based on features that have well-understood medical and physical interpretation. However, classifiers based on morphological features commonly underperform in the presence of the shadowing artifact and ill-defined mass borders, while texture based classifiers may fail when the US image is too noisy. Therefore, in practice it would be beneficial to select the classification method based on the appearance of the particular US image. In this work, we develop a deep meta-network that can automatically process input breast mass US images and recommend whether to apply the shape or texture based classifier for the breast mass differentiation. Our preliminary results demonstrate that meta-learning techniques can be used to improve the performance of the standard classifiers based on handcrafted features. With the proposed meta-learning based approach, we achieved the area under the receiver operating characteristic curve of 0.95 and accuracy of 0.91.
Parameter estimation of the homodyned K distribution based on neural networks and trainable fractional-order moments
Oct 11, 2022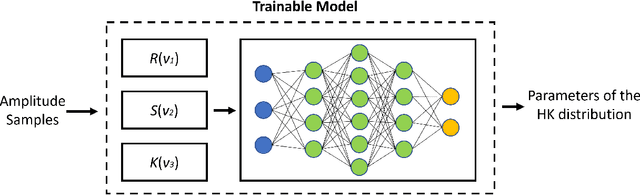
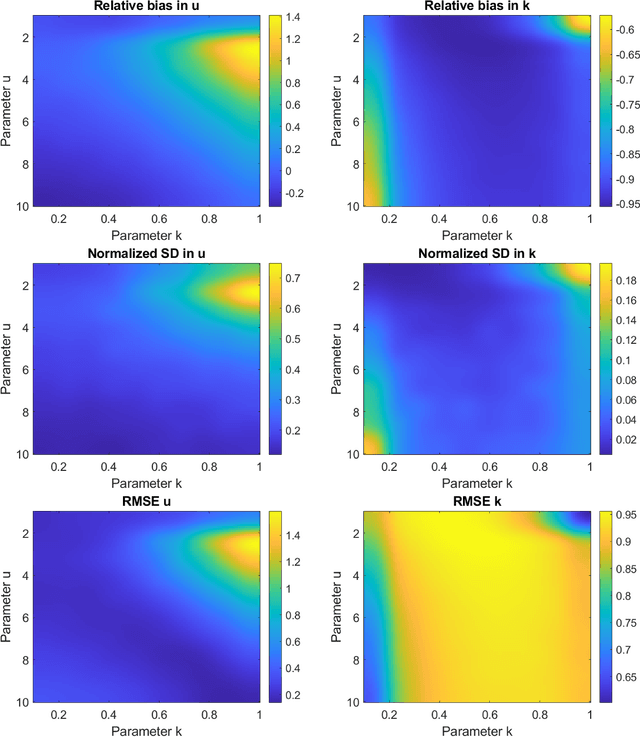
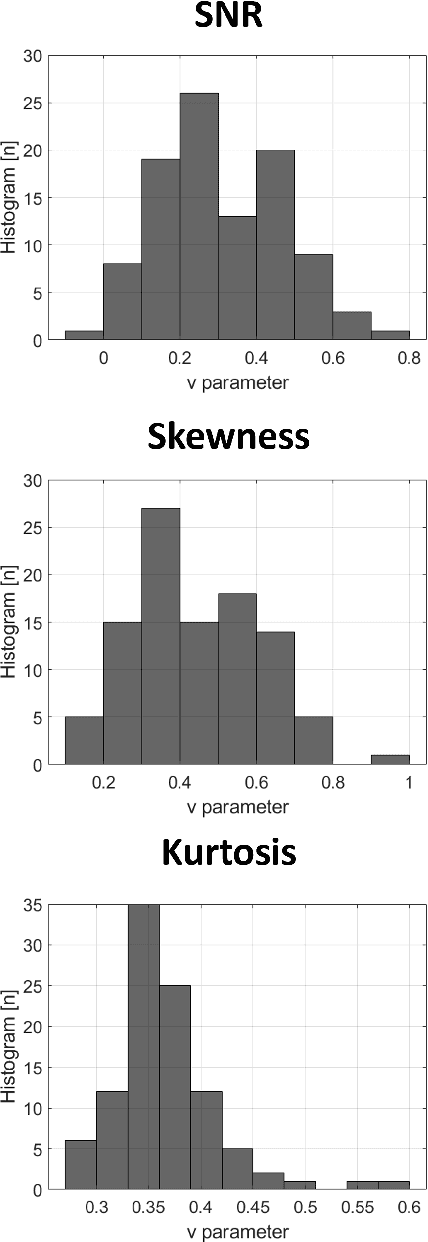
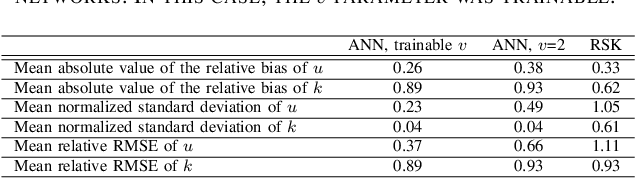
Abstract:Homodyned K (HK) distribution has been widely used to describe the scattering phenomena arising in various research fields, such as ultrasound imaging or optics. In this work, we propose a machine learning based approach to the estimation of the HK distribution parameters. We develop neural networks that can estimate the HK distribution parameters based on the signal-to-noise ratio, skewness and kurtosis calculated using fractional-order moments. Compared to the previous approaches, we consider the orders of the moments as trainable variables that can be optimized along with the network weights using the back-propagation algorithm. Networks are trained based on samples generated from the HK distribution. Obtained results demonstrate that the proposed method can be used to accurately estimate the HK distribution parameters.
Estimating the ultrasound attenuation coefficient using convolutional neural networks -- a feasibility study
May 19, 2022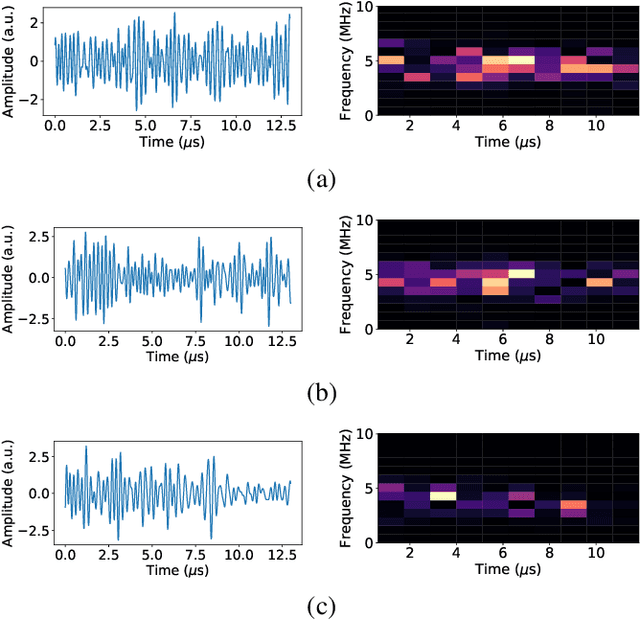
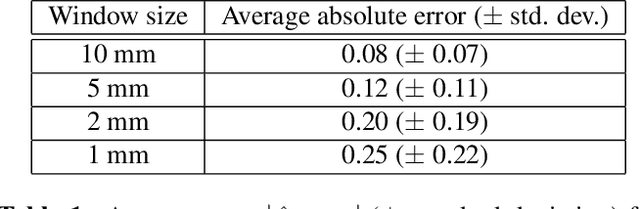
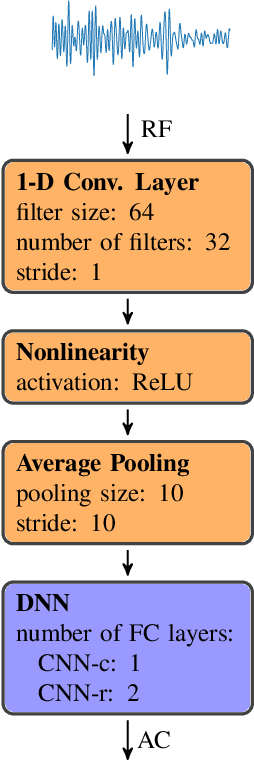
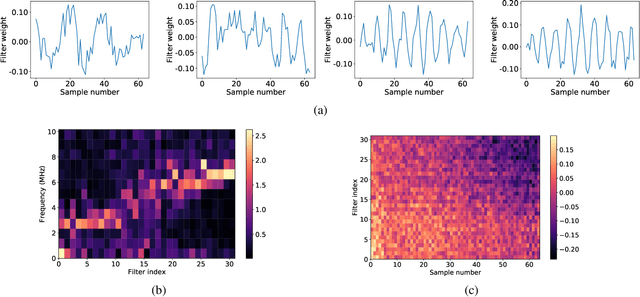
Abstract:Attenuation coefficient (AC) is a fundamental measure of tissue acoustical properties, which can be used in medical diagnostics. In this work, we investigate the feasibility of using convolutional neural networks (CNNs) to directly estimate AC from radio-frequency (RF) ultrasound signals. To develop the CNNs we used RF signals collected from tissue mimicking numerical phantoms for the AC values in a range from 0.1 to 1.5 dB/(MHz*cm). The models were trained based on 1-D patches of RF data. We obtained mean absolute AC estimation errors of 0.08, 0.12, 0.20, 0.25 for the patch lengths: 10 mm, 5 mm, 2 mm and 1 mm, respectively. We explain the performance of the model by visualizing the frequency content associated with convolutional filters. Our study presents that the AC can be calculated using deep learning, and the weights of the CNNs can have physical interpretation.
Breast mass segmentation based on ultrasonic entropy maps and attention gated U-Net
Jan 27, 2020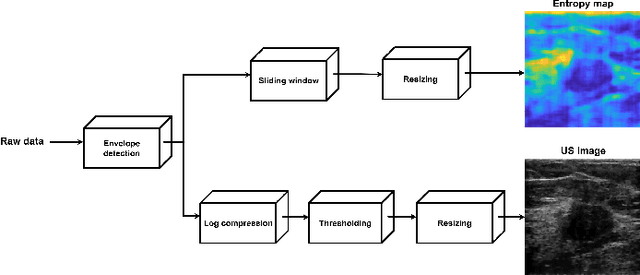
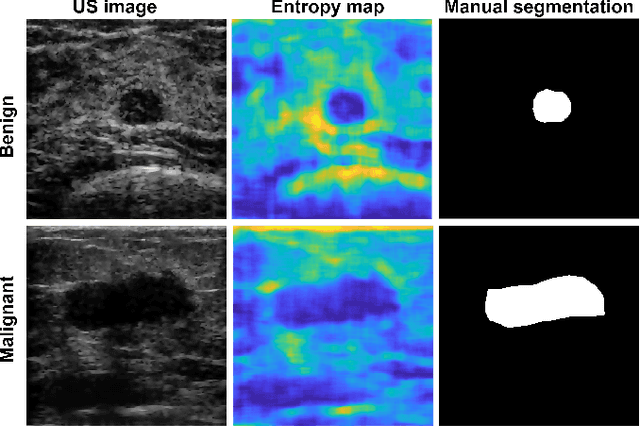
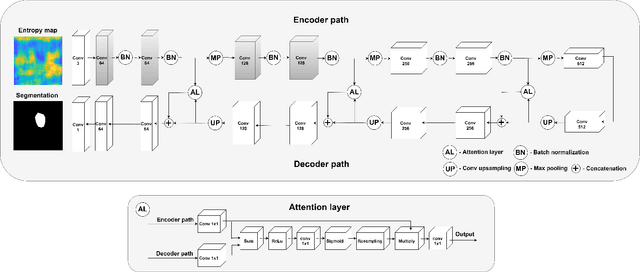
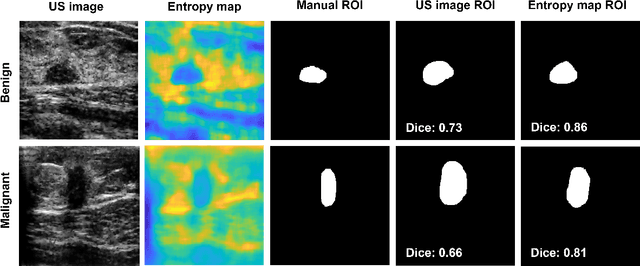
Abstract:We propose a novel deep learning based approach to breast mass segmentation in ultrasound (US) imaging. In comparison to commonly applied segmentation methods, which use US images, our approach is based on quantitative entropy parametric maps. To segment the breast masses we utilized an attention gated U-Net convolutional neural network. US images and entropy maps were generated based on raw US signals collected from 269 breast masses. The segmentation networks were developed separately using US image and entropy maps, and evaluated on a test set of 81 breast masses. The attention U-Net trained based on entropy maps achieved average Dice score of 0.60 (median 0.71), while for the model trained using US images we obtained average Dice score of 0.53 (median 0.59). Our work presents the feasibility of using quantitative US parametric maps for the breast mass segmentation. The obtained results suggest that US parametric maps, which provide the information about local tissue scattering properties, might be more suitable for the development of breast mass segmentation methods than regular US images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge