Katarzyna Dobruch-Sobczak
Breast mass segmentation based on ultrasonic entropy maps and attention gated U-Net
Jan 27, 2020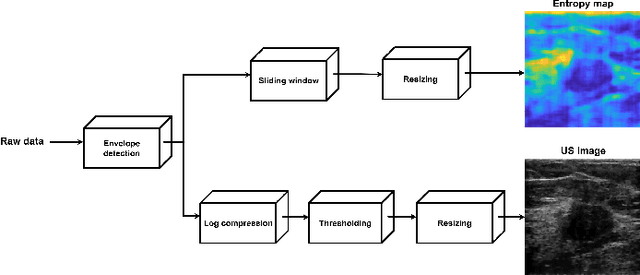
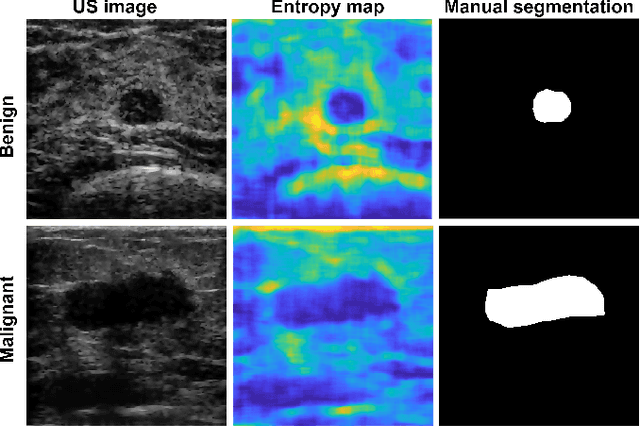
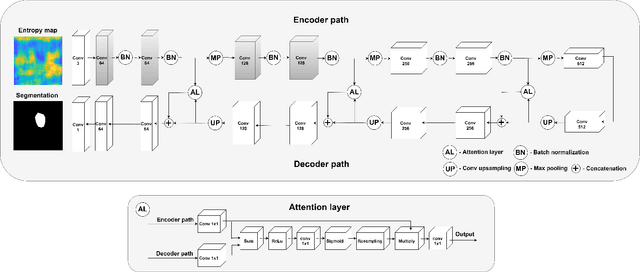
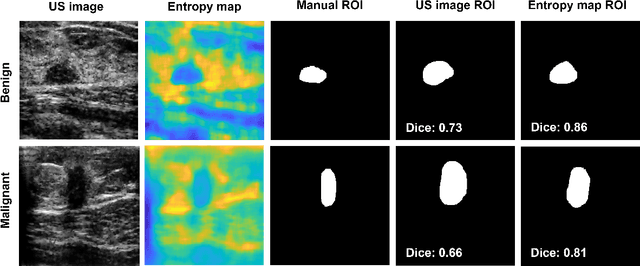
Abstract:We propose a novel deep learning based approach to breast mass segmentation in ultrasound (US) imaging. In comparison to commonly applied segmentation methods, which use US images, our approach is based on quantitative entropy parametric maps. To segment the breast masses we utilized an attention gated U-Net convolutional neural network. US images and entropy maps were generated based on raw US signals collected from 269 breast masses. The segmentation networks were developed separately using US image and entropy maps, and evaluated on a test set of 81 breast masses. The attention U-Net trained based on entropy maps achieved average Dice score of 0.60 (median 0.71), while for the model trained using US images we obtained average Dice score of 0.53 (median 0.59). Our work presents the feasibility of using quantitative US parametric maps for the breast mass segmentation. The obtained results suggest that US parametric maps, which provide the information about local tissue scattering properties, might be more suitable for the development of breast mass segmentation methods than regular US images.
Impact of ultrasound image reconstruction method on breast lesion classification with neural transfer learning
Apr 06, 2018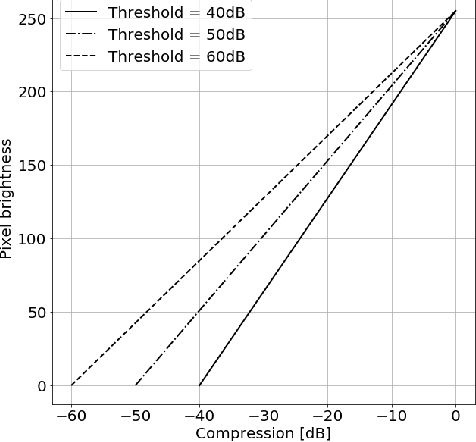

Abstract:Deep learning algorithms, especially convolutional neural networks, have become a methodology of choice in medical image analysis. However, recent studies in computer vision show that even a small modification of input image intensities may cause a deep learning model to classify the image differently. In medical imaging, the distribution of image intensities is related to applied image reconstruction algorithm. In this paper we investigate the impact of ultrasound image reconstruction method on breast lesion classification with neural transfer learning. Due to high dynamic range raw ultrasonic signals are commonly compressed in order to reconstruct B-mode images. Based on raw data acquired from breast lesions, we reconstruct B-mode images using different compression levels. Next, transfer learning is applied for classification. Differently reconstructed images are employed for training and evaluation. We show that the modification of the reconstruction algorithm leads to decrease of classification performance. As a remedy, we propose a method of data augmentation. We show that the augmentation of the training set with differently reconstructed B-mode images leads to a more robust and efficient classification. Our study suggests that it is important to take into account image reconstruction algorithms implemented in medical scanners during development of computer aided diagnosis systems.
Added value of morphological features to breast lesion diagnosis in ultrasound
Jun 06, 2017
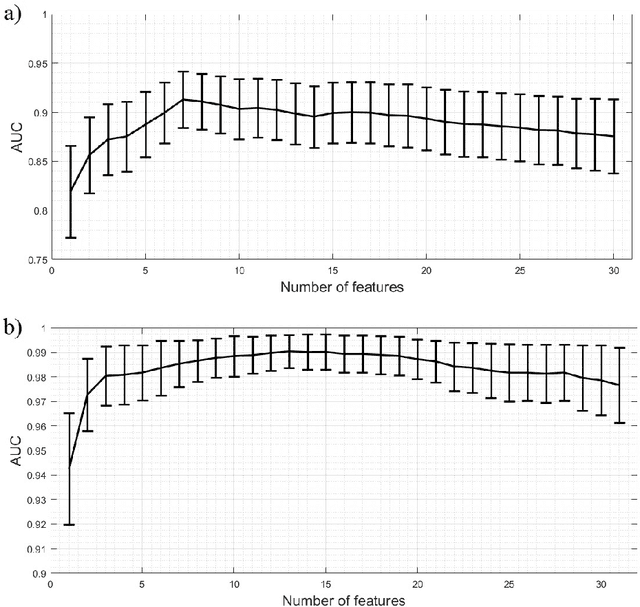
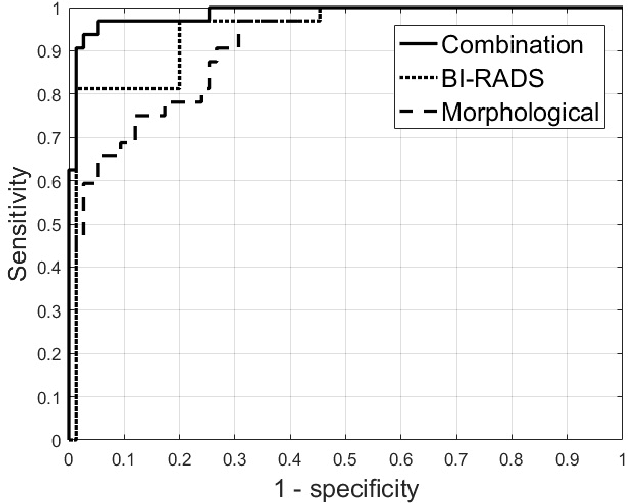
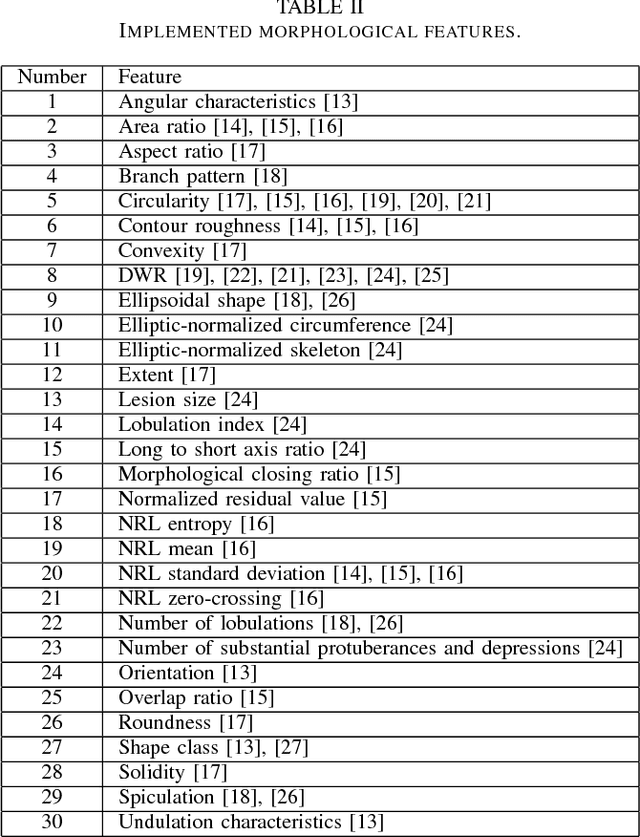
Abstract:Ultrasound imaging plays an important role in breast lesion differentiation. However, diagnostic accuracy depends on ultrasonographer experience. Various computer aided diagnosis systems has been developed to improve breast cancer detection and reduce the number of unnecessary biopsies. In this study, our aim was to improve breast lesion classification based on the BI-RADS (Breast Imaging - Reporting and Data System). This was accomplished by combining the BI-RADS with morphological features which assess lesion boundary. A dataset of 214 lesion images was used for analysis. 30 morphological features were extracted and feature selection scheme was applied to find features which improve the BI-RADS classification performance. Additionally, the best performing morphological feature subset was indicated. We obtained a better classification by combining the BI-RADS with six morphological features. These features were the extent, overlap ratio, NRL entropy, circularity, elliptic-normalized circumference and the normalized residual value. The area under the receiver operating curve calculated with the use of the combined classifier was 0.986. The best performing morphological feature subset contained six features: the DWR, NRL entropy, normalized residual value, overlap ratio, extent and the morphological closing ratio. For this set, the area under the curve was 0.901. The combination of the radiologist's experience related to the BI-RADS and the morphological features leads to a more effective breast lesion classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge